Network-Guided GWAS Improves Identification of Genes Affecting Free Amino Acids
- PMID: 27872244
- PMCID: PMC5210728
- DOI: 10.1104/pp.16.01287
Network-Guided GWAS Improves Identification of Genes Affecting Free Amino Acids
Abstract
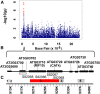
Amino acids are essential for proper growth and development in plants. Amino acids serve as building blocks for proteins but also are important for responses to stress and the biosynthesis of numerous essential compounds. In seed, the pool of free amino acids (FAAs) also contributes to alternative energy, desiccation, and seed vigor; thus, manipulating FAA levels can significantly impact a seed's nutritional qualities. While genome-wide association studies (GWAS) on branched-chain amino acids have identified some regulatory genes controlling seed FAAs, the genetic regulation of FAA levels, composition, and homeostasis in seeds remains mostly unresolved. Hence, we performed GWAS on 18 FAAs from a 313-ecotype Arabidopsis (Arabidopsis thaliana) association panel. Specifically, GWAS was performed on 98 traits derived from known amino acid metabolic pathways (approach 1) and then on 92 traits generated from an unbiased correlation-based metabolic network analysis (approach 2), and the results were compared. The latter approach facilitated the discovery of additional novel metabolic interactions and single-nucleotide polymorphism-trait associations not identified by the former approach. The most prominent network-guided GWAS signal was for a histidine (His)-related trait in a region containing two genes: a cationic amino acid transporter (CAT4) and a polynucleotide phosphorylase resistant to inhibition with fosmidomycin. A reverse genetics approach confirmed CAT4 to be responsible for the natural variation of His-related traits across the association panel. Given that His is a semiessential amino acid and a potent metal chelator, CAT4 orthologs could be considered as candidate genes for seed quality biofortification in crop plants.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures




References
-
- Angelovici R, Fait A, Fernie AR, Galili G (2011) A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol 189: 148–159 - PubMed
-
- Angelovici R, Galili G, Fernie AR, Fait A (2010) Seed desiccation: a bridge between maturation and germination. Trends Plant Sci 15: 211–218 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

