Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA
- PMID: 27872306
- PMCID: PMC5150408
- DOI: 10.1073/pnas.1617789113
Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA
Erratum in
-
Correction for Lorenzi et al., Paired quantitative and qualitative assessment of the replication-competent HIV-1 reservoir and comparison with integrated proviral DNA.Proc Natl Acad Sci U S A. 2017 Jan 24;114(4):E648-E649. doi: 10.1073/pnas.1619801114. Epub 2017 Jan 17. Proc Natl Acad Sci U S A. 2017. PMID: 28096398 Free PMC article. No abstract available.
Abstract
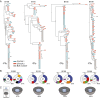
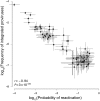
HIV-1-infected individuals harbor a latent reservoir of infected CD4+ T cells that is not eradicated by antiretroviral therapy (ART). This reservoir presents the greatest barrier to an HIV-1 cure and has remained difficult to characterize, in part, because the vast majority of integrated sequences are defective and incapable of reactivation. To characterize the replication-competent reservoir, we have combined two techniques, quantitative viral outgrowth and qualitative sequence analysis of clonal outgrowth viruses. Leukapheresis samples from four fully ART-suppressed, chronically infected individuals were assayed at two time points separated by a 4- to 6-mo interval. Overall, 54% of the viruses emerging from the latent reservoir showed gp160 env sequences that were identical to at least one other virus. Moreover, 43% of the env sequences from viruses emerging from the reservoir were part of identical groups at the two time points. Groups of identical expanded sequences made up 54% of proviral DNA, and, as might be expected, the sequences of replication-competent viruses in the active reservoir showed limited overlap with integrated proviral DNA, most of which is known to represent defective viruses. Finally, there was an inverse correlation between proviral DNA clone size and the probability of reactivation, suggesting that replication-competent viruses are less likely to be found among highly expanded provirus-containing cell clones.
Keywords: HIV; culture; method; replication-competent; reservoir.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







References
-
- Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–517. - PubMed
-
- Churchill MJ, Deeks SG, Margolis DM, Siliciano RF, Swanstrom R. HIV reservoirs: What, where and how to target them. Nat Rev Microbiol. 2016;14(1):55–60. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

