Identification of Polo-like kinase 1 interaction inhibitors using a novel cell-based assay
- PMID: 27874094
- PMCID: PMC5118709
- DOI: 10.1038/srep37581
Identification of Polo-like kinase 1 interaction inhibitors using a novel cell-based assay
Abstract
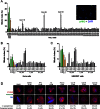
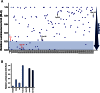
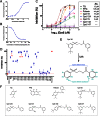
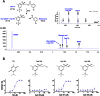
Polo-like kinase 1 (Plk1) plays several roles in cell division and it is a recognized cancer drug target. Plk1 levels are elevated in cancer and several types of cancer cells are hypersensitive to Plk1 inhibition. Small molecule inhibitors of the kinase domain (KD) of Plk1 have been developed. Their selectivity is limited, which likely contributes to their toxicity. Polo-like kinases are characterized by a Polo-Box Domain (PBD), which mediates interactions with phosphorylation substrates or regulators. Inhibition of the PBD could allow better selectivity or result in different effects than inhibition of the KD. In vitro screens have been used to identify PBD inhibitors with mixed results. We developed the first cell-based assay to screen for PBD inhibitors, using Bioluminescence Resonance Energy Transfer (BRET). We screened through 112 983 compounds and characterized hits in secondary biochemical and biological assays. Subsequent Structure-Activity Relationship (SAR) analysis on our most promising hit revealed that it requires an alkylating function for its activity. In addition, we show that the previously reported PBD inhibitors thymoquinone and Poloxin are also alkylating agents. Our cell-based assay is a promising tool for the identification of new PBD inhibitors with more drug-like profiles using larger and more diverse chemical libraries.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

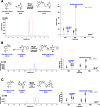


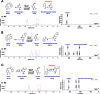
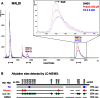
References
-
- Llamazares S. et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev 5, 2153–65 (1991). - PubMed
-
- Sunkel C. E. & Glover D. M. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci 89 (Pt 1), 25–38 (1988). - PubMed
-
- Archambault V. & Glover D. M. Polo-like kinases: conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol 10, 265–75 (2009). - PubMed
-
- Elia A. E., Cantley L. C. & Yaffe M. B. Proteomic screen finds pSer/pThr-binding domain localizing Plk1 to mitotic substrates. Science 299, 1228–31 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

