Family-wide Structural Characterization and Genomic Comparisons Decode the Diversity-oriented Biosynthesis of Thalassospiramides by Marine Proteobacteria
- PMID: 27875306
- PMCID: PMC5207150
- DOI: 10.1074/jbc.M116.756858
Family-wide Structural Characterization and Genomic Comparisons Decode the Diversity-oriented Biosynthesis of Thalassospiramides by Marine Proteobacteria
Abstract
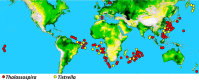
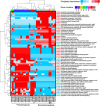
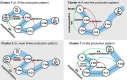
The thalassospiramide lipopeptides have great potential for therapeutic applications; however, their structural and functional diversity and biosynthesis are poorly understood. Here, by cultivating 130 Rhodospirillaceae strains sampled from oceans worldwide, we discovered 21 new thalassospiramide analogues and demonstrated their neuroprotective effects. To investigate the diversity of biosynthetic gene cluster (BGC) architectures, we sequenced the draft genomes of 28 Rhodospirillaceae strains. Our family-wide genomic analysis revealed three types of dysfunctional BGCs and four functional BGCs whose architectures correspond to four production patterns. This correlation allowed us to reassess the "diversity-oriented biosynthesis" proposed for the microbial production of thalassospiramides, which involves iteration of several key modules. Preliminary evolutionary investigation suggested that the functional BGCs could have arisen through module/domain loss, whereas the dysfunctional BGCs arose through horizontal gene transfer. Further comparative genomics indicated that thalassospiramide production is likely to be attendant on particular genes/pathways for amino acid metabolism, signaling transduction, and compound efflux. Our findings provide a systematic understanding of thalassospiramide production and new insights into the underlying mechanism.
Keywords: bacteria; biosynthesis; genomics; natural product; neuroprotection.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Investigating the Biosynthesis of Natural Products from Marine Proteobacteria: A Survey of Molecules and Strategies.Mar Drugs. 2017 Aug 1;15(8):235. doi: 10.3390/md15080235. Mar Drugs. 2017. PMID: 28762997 Free PMC article. Review.
-
Biosynthetic multitasking facilitates thalassospiramide structural diversity in marine bacteria.J Am Chem Soc. 2013 Jan 23;135(3):1155-62. doi: 10.1021/ja3119674. Epub 2013 Jan 11. J Am Chem Soc. 2013. PMID: 23270364 Free PMC article.
-
Genomic insights into the evolution of hybrid isoprenoid biosynthetic gene clusters in the MAR4 marine streptomycete clade.BMC Genomics. 2015 Nov 17;16:960. doi: 10.1186/s12864-015-2110-3. BMC Genomics. 2015. PMID: 26578069 Free PMC article.
-
Thalassospiramide G, a new γ-amino-acid-bearing peptide from the marine bacterium Thalassospira sp.Mar Drugs. 2013 Feb 26;11(3):611-22. doi: 10.3390/md11030611. Mar Drugs. 2013. PMID: 23442790 Free PMC article.
-
Natural Products and the Gene Cluster Revolution.Trends Microbiol. 2016 Dec;24(12):968-977. doi: 10.1016/j.tim.2016.07.006. Epub 2016 Aug 1. Trends Microbiol. 2016. PMID: 27491886 Free PMC article. Review.
Cited by
-
Neuroinflammation in the Central Nervous System: Exploring the Evolving Influence of Endocannabinoid System.Biomedicines. 2023 Sep 26;11(10):2642. doi: 10.3390/biomedicines11102642. Biomedicines. 2023. PMID: 37893016 Free PMC article. Review.
-
Pass-back chain extension expands multimodular assembly line biosynthesis.Nat Chem Biol. 2020 Jan;16(1):42-49. doi: 10.1038/s41589-019-0385-4. Epub 2019 Oct 21. Nat Chem Biol. 2020. PMID: 31636431 Free PMC article.
-
Investigating the Biosynthesis of Natural Products from Marine Proteobacteria: A Survey of Molecules and Strategies.Mar Drugs. 2017 Aug 1;15(8):235. doi: 10.3390/md15080235. Mar Drugs. 2017. PMID: 28762997 Free PMC article. Review.
-
Industrial and Pharmaceutical Applications of Microbial Diversity of Hypersaline Ecology from Lonar Soda Crater.Curr Pharm Biotechnol. 2024;25(12):1564-1584. doi: 10.2174/0113892010265978231109085224. Curr Pharm Biotechnol. 2024. PMID: 38258768 Review.
-
Targeting eukaryotic proteases for natural products-based drug development.Nat Prod Rep. 2020 Jun 24;37(6):827-860. doi: 10.1039/c9np00060g. Nat Prod Rep. 2020. PMID: 32519686 Free PMC article. Review.
References
-
- Cotter P. D., Hill C., and Ross R. P. (2005) Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3, 777–788 - PubMed
-
- Oh D. C., Strangman W. K., Kauffman C. A., Jensen P. R., and Fenical W. (2007) Thalassospiramides A and B, immunosuppressive peptides from the marine bacterium Thalassospira sp. Org. Lett. 9, 1525–1528 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

