Engineering an inducible gene expression system for Bacillus subtilis from a strong constitutive promoter and a theophylline-activated synthetic riboswitch
- PMID: 27876054
- PMCID: PMC5120567
- DOI: 10.1186/s12934-016-0599-z
Engineering an inducible gene expression system for Bacillus subtilis from a strong constitutive promoter and a theophylline-activated synthetic riboswitch
Abstract
Background: Synthetic riboswitches have been increasingly used to control and tune gene expression in diverse organisms. Although a set of theophylline-responsive riboswitches have been developed for bacteria, fully functional expression elements mediated by synthetic riboswitches in Bacillus subtilis are rarely used because of the host-dependent compatibility between the promoters and riboswitches.
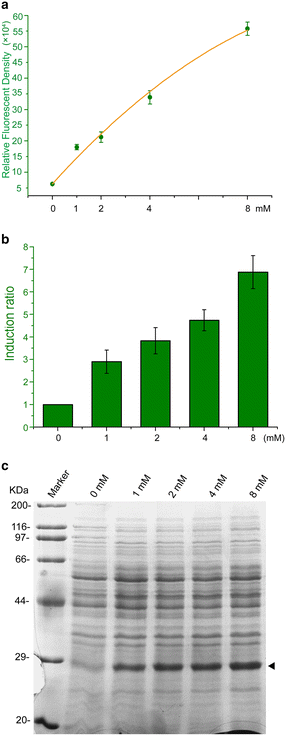
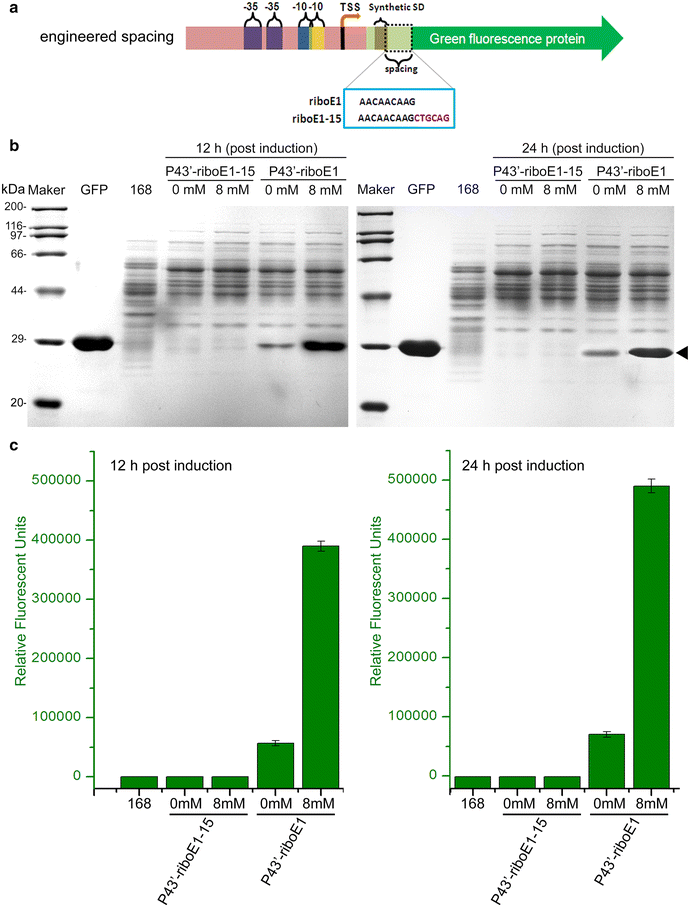
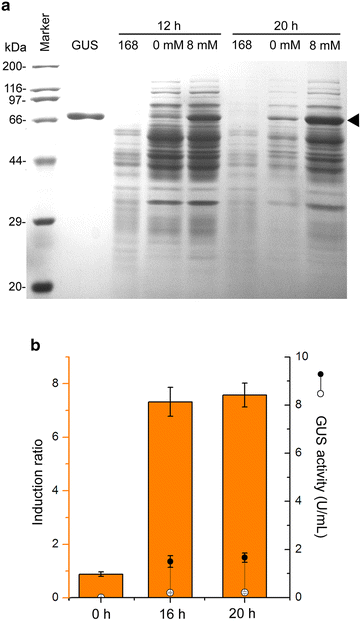
Results: A novel genetic element composed of the promoter P43 and a theophylline-riboswitch was developed and characterized in B. subtilis. When combined with a P43 promoter (P43'-riboE1), the theophylline-riboswitch successfully switched the constitutive expression pattern of P43 to an induced pattern. The expression mediated by the novel element could be activated at the translational level by theophylline with a relatively high induction ratio. The induction ratios for P43'-riboE1 by 4-mM theophylline were elevated during the induction period. The level of induced expression was dependent on the theophylline dose. Correspondingly, the induction ratios gradually increased in parallel with the elevated dose of theophylline. Importantly, the induced expression level was higher than three other strong constitutive promoters including PsrfA, PaprE, and the native P43. It was found that the distance between the SD sequence within the expression element and the start codon significantly influenced both the level of induced expression and the induction ratio. A 9-bp spacer was suitable for producing desirable expression level and induction ratio. Longer spacer reduced the activation efficiency. Importantly, the system successfully overexpressed β-glucuronidase at equal levels, and induction ratio was similar to that of GFP.
Conclusion: The constructed theophylline-inducible gene expression system has broad compatibility and robustness, which has great potential in over-production of pharmaceutical and industrial proteins and utilization in building more complex gene circuits.
Keywords: Bacillus subtilis; Controllable gene expression; RNA switch; Synthetic riboswitches; Theophylline.
Figures



Similar articles
-
Comprehensive characterization of a theophylline riboswitch reveals two pivotal features of Shine-Dalgarno influencing activated translation property.Appl Microbiol Biotechnol. 2017 Mar;101(5):2107-2120. doi: 10.1007/s00253-016-7988-4. Epub 2016 Dec 16. Appl Microbiol Biotechnol. 2017. PMID: 27986992
-
[Construction and application of theophylline-activated RNA switches in the regulation of expression of recombinant proteins in Bacillus subtilis].Sheng Wu Gong Cheng Xue Bao. 2019 Aug 25;35(8):1478-1490. doi: 10.13345/j.cjb.190037. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31441619 Chinese.
-
Development of a novel strategy for robust synthetic bacterial promoters based on a stepwise evolution targeting the spacer region of the core promoter in Bacillus subtilis.Microb Cell Fact. 2019 May 29;18(1):96. doi: 10.1186/s12934-019-1148-3. Microb Cell Fact. 2019. PMID: 31142347 Free PMC article.
-
Exploitation of Bacillus subtilis as a robust workhorse for production of heterologous proteins and beyond.World J Microbiol Biotechnol. 2018 Sep 10;34(10):145. doi: 10.1007/s11274-018-2531-7. World J Microbiol Biotechnol. 2018. PMID: 30203131 Review.
-
Engineering and In Vivo Applications of Riboswitches.Annu Rev Biochem. 2017 Jun 20;86:515-539. doi: 10.1146/annurev-biochem-060815-014628. Epub 2017 Mar 30. Annu Rev Biochem. 2017. PMID: 28375743 Review.
Cited by
-
Recent trends in biocatalysis.Chem Soc Rev. 2021 Jul 21;50(14):8003-8049. doi: 10.1039/d0cs01575j. Epub 2021 Jun 18. Chem Soc Rev. 2021. PMID: 34142684 Free PMC article. Review.
-
Using the IPTG-Inducible Pgrac212 Promoter for Overexpression of Human Rhinovirus 3C Protease Fusions in the Cytoplasm of Bacillus subtilis Cells.Curr Microbiol. 2019 Dec;76(12):1477-1486. doi: 10.1007/s00284-019-01783-9. Epub 2019 Oct 14. Curr Microbiol. 2019. PMID: 31612259
-
An overview and future prospects of recombinant protein production in Bacillus subtilis.Appl Microbiol Biotechnol. 2021 Sep;105(18):6607-6626. doi: 10.1007/s00253-021-11533-2. Epub 2021 Sep 1. Appl Microbiol Biotechnol. 2021. PMID: 34468804 Review.
-
The modular pYT vector series employed for chromosomal gene integration and expression to produce carbazoles and glycolipids in P. putida.FEMS Microbes. 2022 Dec 19;4:xtac030. doi: 10.1093/femsmc/xtac030. eCollection 2023. FEMS Microbes. 2022. PMID: 37333445 Free PMC article.
-
Influence of N-terminal His-tags on the production of recombinant proteins in the cytoplasm of Bacillus subtilis.Biotechnol Rep (Amst). 2022 Jul 19;35:e00754. doi: 10.1016/j.btre.2022.e00754. eCollection 2022 Sep. Biotechnol Rep (Amst). 2022. PMID: 35911505 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

