A framework for the detection of de novo mutations in family-based sequencing data
- PMID: 27876817
- PMCID: PMC5255947
- DOI: 10.1038/ejhg.2016.147
A framework for the detection of de novo mutations in family-based sequencing data
Abstract
Germline mutation detection from human DNA sequence data is challenging due to the rarity of such events relative to the intrinsic error rates of sequencing technologies and the uneven coverage across the genome. We developed PhaseByTransmission (PBT) to identify de novo single nucleotide variants and short insertions and deletions (indels) from sequence data collected in parent-offspring trios. We compute the joint probability of the data given the genotype likelihoods in the individual family members, the known familial relationships and a prior probability for the mutation rate. Candidate de novo mutations (DNMs) are reported along with their posterior probability, providing a systematic way to prioritize them for validation. Our tool is integrated in the Genome Analysis Toolkit and can be used together with the ReadBackedPhasing module to infer the parental origin of DNMs based on phase-informative reads. Using simulated data, we show that PBT outperforms existing tools, especially in low coverage data and on the X chromosome. We further show that PBT displays high validation rates on empirical parent-offspring sequencing data for whole-exome data from 104 trios and X-chromosome data from 249 parent-offspring families. Finally, we demonstrate an association between father's age at conception and the number of DNMs in female offspring's X chromosome, consistent with previous literature reports.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
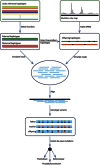


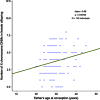
References
-
- Genome of the Netherlands Consortium: Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet 2014; 46: 818–825. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

