Forward Genetic Screen in Caenorhabditis elegans Suggests F57A10.2 and acp-4 As Suppressors of C9ORF72 Related Phenotypes
- PMID: 27877110
- PMCID: PMC5100550
- DOI: 10.3389/fnmol.2016.00113
Forward Genetic Screen in Caenorhabditis elegans Suggests F57A10.2 and acp-4 As Suppressors of C9ORF72 Related Phenotypes
Abstract
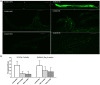
An abnormally expanded GGGGCC repeat in C9ORF72 is the most frequent causal mutation associated with amyotrophic lateral sclerosis (ALS)/frontotemporal lobar degeneration (FTLD). Both gain-of-function (gf) and loss-of-function (lf) mechanisms have been involved in C9ORF72 related ALS/FTLD. The gf mechanism of C9ORF72 has been studied in various animal models but not in C. elegans. In the present study, we described mutant C9ORF72 modeling in C. elegans and report the finding of two suppressor genes. We made transgenes containing 9 or 29 repeats of GGGGCC in C9ORF72, driven by either the hsp-16 promoters or the unc-119 promoter. Transgenic worms were made to carry such transgenes. Phenotypic analysis of those animals revealed that Phsp-16::(G4C2)29::GFP transgenic animals (EAB 135) displayed severe paralysis by the second day of adulthood, followed by lethality, which phenotypes were less severe in Phsp-16::(G4C2)9::GFP transgenic animals (EAB242), and absent in control strains expressing empty vectors. Suppressor genes of this locomotor phenotype were pursued by introducing mutations with ethyl methanesulfonate in EAB135, screening mutant strains that moved faster than EAB135 by a food-ring assay, identifying mutations by whole-genome sequencing and testing the underlying mechanism of the suppressor genes either by employing RNA interference studies or C. elegans genetics. Three mutant strains, EAB164, EAB165 and EAB167, were identified. Eight suppressor genes carrying nonsense/canonical splicing site mutations were confirmed, among which a nonsense mutation of F57A10.2/VAMP was found in all three mutant strains, and a nonsense mutation of acp-4/ACP2 was only found in EAB164. Knock down/out of those two genes in EAB135 animals by feeding RNAi/introducing a known acp-4 null allele phenocopied the suppression of the C9ORF72 variant related movement defect in the mutant strains. Translational conformation in a mammalian system is required, but our worm data suggest that altering acp-4/ACP2 encoding lysosomal acid phosphatase may provide a potential therapeutic method of reducing acp-4/ACP2 levels, as opposed or complementary to directly reducing C9ORF72, to relieve C9ORF72-ALS phenotypes. It also suggests that the C9ORF72-ALS/FTLD may share a pathophysiologic mechanism with vesicle-associated membrane protein-associated protein B, a homolog of F57A10.2/VAMP, which is a proven ALS8 gene.
Keywords: ACP2; ALS-FTLD; C9ORF72; Caenorhabditis elegans; VAPB; amyotrophic lateral sclerosis.
Figures


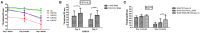
References
-
- Baker D. J., Blackburn D. J., Keatinge M., Sokhi D., Viskaitis P., Heath P. R., et al. (2015). Lysosomal and phagocytic activity is increased in astrocytes during disease progression in the SOD1 (G93A) mouse model of amyotrophic lateral sclerosis. Front. Cell. Neurosci. 9:410. 10.3389/fncel.2015.00410 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

