Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation
- PMID: 27878953
- PMCID: PMC5387161
- DOI: 10.1111/jcmm.13035
Long non-coding RNA linc00261 suppresses gastric cancer progression via promoting Slug degradation
Abstract
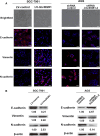
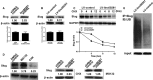
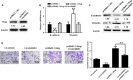
Gastric cancer (GC) remains a threat to public health with high incidence and mortality worldwide. Increasing evidence demonstrates that long non-coding RNAs (lncRNAs) play critical regulatory roles in cancer biology, including GC. Previous profiling study showed that lncRNA linc00261 was aberrantly expressed in GC. However, the role of linc00261 in GC progression and the precise molecular mechanism remain unknown. In this study, we report that linc00261 was significantly down-regulated in GC tissues and the expression level of linc00261 negatively correlated with advanced tumour status and clinical stage as well as poor prognostic outcome. In vitro functional assays indicate that ectopic expression of linc00261 suppressed cell invasion by inhibiting the epithelial-mesenchymal transition (EMT). By RNA pull-down and mass spectrum experiments, we identified Slug as an RNA-binding protein that binds to linc00261. We confirmed that linc00261 down-regulated Slug by decreasing the stability of Slug proteins and that the tumour-suppressive function of linc00261 can be neutralized by Slug. linc00261 may promote the degradation of Slug via enhancing the interaction between GSK3β and Slug. Moreover, linc00216 overexpression repressed lung metastasis in vivo. Together, our findings suggest that linc00261 acts a tumour suppressor in GC by decreasing the stability of Slug proteins and suppressing EMT. By clarifying the mechanisms underlying GC progression, these findings may facilitate the development of novel therapeutic strategies for GC.
Keywords: Slug; epithelial-mesenchymal transition; gastric cancer; invasion; long non-coding RNA.
© 2016 The Authors. Journal of Cellular and Molecular Medicine published by John Wiley & Sons Ltd and Foundation for Cellular and Molecular Medicine.
Figures






References
-
- Ferro A, Peleteiro B, Malvezzi M, et al Worldwide trends in gastric cancer mortality (1980–2011), with predictions to 2015, and incidence by subtype. Eur J Cancer. 2014; 50: 1330–44. - PubMed
-
- Dassen AE, Dikken JL, van de Velde CJ, et al Changes in treatment patterns and their influence on long‐term survival in patients with stages I‐III gastric cancer in The Netherlands. Int J Cancer. 2013; 133: 1859–66. - PubMed
-
- Zang ZJ, Cutcutache I, Poon SL, et al Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012; 44: 570–4. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

