Differential expression of cucumber RNA-dependent RNA polymerase 1 genes during antiviral defence and resistance
- PMID: 27879040
- PMCID: PMC6637986
- DOI: 10.1111/mpp.12518
Differential expression of cucumber RNA-dependent RNA polymerase 1 genes during antiviral defence and resistance
Abstract
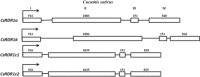
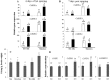
RNA-dependent RNA polymerase 1 (RDR1) plays a crucial role in plant defence against viruses. In this study, it was observed that cucumber, Cucumis sativus, uniquely encodes a small gene family of four RDR1 genes. The cucumber RDR1 genes (CsRDR1a, CsRDR1b and duplicated CsRDR1c1/c2) shared 55%-60% homology in their encoded amino acid sequences. In healthy cucumber plants, RDR1a and RDR1b transcripts were expressed at higher levels than transcripts of RDR1c1/c2, which were barely detectable. The expression of all four CsRDR1 genes was induced by virus infection, after which the expression level of CsRDR1b increased 10-20-fold in several virus-resistant cucumber cultivars and in a broad virus-resistant transgenic cucumber line expressing a high level of transgene small RNAs, all without alteration in salicylic acid (SA) levels. By comparison, CsRDR1c1/c2 genes were highly induced (25-1300-fold) in susceptible cucumber cultivars infected with RNA or DNA viruses. Inhibition of RDR1c1/c2 expression led to increased virus accumulation. Ectopic application of SA induced the expression of cucumber RDR1a, RDR1b and RDRc1/c2 genes. A constitutive high level of RDR1b gene expression independent of SA was found to be associated with broad virus resistance. These findings show that multiple RDR1 genes are involved in virus resistance in cucumber and are regulated in a coordinated fashion with different expression profiles.
Keywords: RDR1; cucumber; gene expression; silencing; virus resistance.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Figures






Similar articles
-
Cucumber RDR1s and cucumber mosaic virus suppressor protein 2b association directs host defence in cucumber plants.Mol Plant Pathol. 2021 Nov;22(11):1317-1331. doi: 10.1111/mpp.13112. Epub 2021 Aug 6. Mol Plant Pathol. 2021. PMID: 34355485 Free PMC article.
-
Analysis of the RNA-Dependent RNA Polymerase 1 (RDR1) Gene Family in Melon.Plants (Basel). 2022 Jul 7;11(14):1795. doi: 10.3390/plants11141795. Plants (Basel). 2022. PMID: 35890429 Free PMC article.
-
RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature.Plant J. 2007 Apr;50(1):40-53. doi: 10.1111/j.1365-313X.2007.03030.x. Epub 2007 Mar 5. Plant J. 2007. PMID: 17346266
-
Antiviral role of plant-encoded RNA-dependent RNA polymerases revisited with deep sequencing of small interfering RNAs of virus origin.Mol Plant Microbe Interact. 2010 Oct;23(10):1248-52. doi: 10.1094/MPMI-06-10-0124. Mol Plant Microbe Interact. 2010. PMID: 20831405 Review.
-
Replicase-mediated resistance: a novel type of virus resistance in transgenic plants?Trends Microbiol. 1994 Feb;2(2):60-3. doi: 10.1016/0966-842x(94)90128-7. Trends Microbiol. 1994. PMID: 8162444 Review.
Cited by
-
Plant A20/AN1 protein serves as the important hub to mediate antiviral immunity.PLoS Pathog. 2018 Sep 13;14(9):e1007288. doi: 10.1371/journal.ppat.1007288. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30212572 Free PMC article.
-
Unravelling cucumber resistance to several viruses via genome-wide association studies highlighted resistance hotspots and new QTLs.Hortic Res. 2022 Aug 25;9:uhac184. doi: 10.1093/hr/uhac184. eCollection 2022. Hortic Res. 2022. PMID: 36338844 Free PMC article.
-
Global Transcriptomic Changes Induced by Infection of Cucumber (Cucumis sativus L.) with Mild and Severe Variants of Hop Stunt Viroid.Front Microbiol. 2017 Dec 12;8:2427. doi: 10.3389/fmicb.2017.02427. eCollection 2017. Front Microbiol. 2017. PMID: 29312160 Free PMC article.
-
Mapping Cucumber Vein Yellowing Virus Resistance in Cucumber (Cucumis sativus L.) by Using BSA-seq Analysis.Front Plant Sci. 2019 Dec 3;10:1583. doi: 10.3389/fpls.2019.01583. eCollection 2019. Front Plant Sci. 2019. PMID: 31850047 Free PMC article.
-
Recovery from virus infection: plant's armory in action.Planta. 2023 Apr 28;257(6):103. doi: 10.1007/s00425-023-04137-9. Planta. 2023. PMID: 37115475 Review.
References
-
- Alamillo, J.M. , Saénz, P. and García, J.A. (2006) Salicylic acid‐mediated and RNA‐silencing defense mechanisms cooperate in the restriction of systemic spread of plum pox virus in tobacco. Plant J. 48, 217–227. - PubMed
-
- Arazi, T. , Slutsky, S.G. , Shiboleth, Y.M. , Wang, Y. , Rubinstein, M. , Barak, S. , Yang, J. and Gal‐On, A. (2001) Engineering zucchini yellow mosaic potyvirus as a non‐pathogenic vector for expression of heterologous proteins in cucurbits. J. Biotechnol. 87, 67–82. - PubMed
-
- Baulcombe, D. (2004) RNA silencing in plants. Nature, 431, 356–363. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

