Phosphorylation of MAP65-1 by Arabidopsis Aurora Kinases Is Required for Efficient Cell Cycle Progression
- PMID: 27879390
- PMCID: PMC5210758
- DOI: 10.1104/pp.16.01602
Phosphorylation of MAP65-1 by Arabidopsis Aurora Kinases Is Required for Efficient Cell Cycle Progression
Abstract
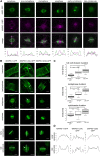
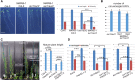
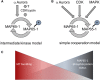
Aurora kinases are key effectors of mitosis. Plant Auroras are functionally divided into two clades. The alpha Auroras (Aurora1 and Aurora2) associate with the spindle and the cell plate and are implicated in controlling formative divisions throughout plant development. The beta Aurora (Aurora3) localizes to centromeres and likely functions in chromosome separation. In contrast to the wealth of data available on the role of Aurora in other kingdoms, knowledge on their function in plants is merely emerging. This is exemplified by the fact that only histone H3 and the plant homolog of TPX2 have been identified as Aurora substrates in plants. Here we provide biochemical, genetic, and cell biological evidence that the microtubule-bundling protein MAP65-1-a member of the MAP65/Ase1/PRC1 protein family, implicated in central spindle formation and cytokinesis in animals, yeasts, and plants-is a genuine substrate of alpha Aurora kinases. MAP65-1 interacts with Aurora1 in vivo and is phosphorylated on two residues at its unfolded tail domain. Its overexpression and down-regulation antagonistically affect the alpha Aurora double mutant phenotypes. Phospho-mutant analysis shows that Aurora contributes to the microtubule bundling capacity of MAP65-1 in concert with other mitotic kinases.
© 2017 American Society of Plant Biologists. All Rights Reserved.
Figures


References
-
- Adams RR, Wheatley SP, Gouldsworthy AM, Kandels-Lewis SE, Carmena M, Smythe C, Gerloff DL, Earnshaw WC (2000) INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol 10: 1075–1078 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

