The Emergence of Pan-Cancer CIMP and Its Elusive Interpretation
- PMID: 27879658
- PMCID: PMC5197955
- DOI: 10.3390/biom6040045
The Emergence of Pan-Cancer CIMP and Its Elusive Interpretation
Abstract
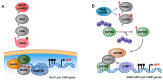
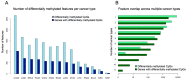
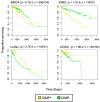
Epigenetic dysregulation is recognized as a hallmark of cancer. In the last 16 years, a CpG island methylator phenotype (CIMP) has been documented in tumors originating from different tissues. However, a looming question in the field is whether or not CIMP is a pan-cancer phenomenon or a tissue-specific event. Here, we give a synopsis of the history of CIMP and describe the pattern of DNA methylation that defines the CIMP phenotype in different cancer types. We highlight new conceptual approaches of classifying tumors based on CIMP in a cancer type-agnostic way that reveal the presence of distinct CIMP tumors in a multitude of The Cancer Genome Atlas (TCGA) datasets, suggesting that this phenotype may transcend tissue-type specificity. Lastly, we show evidence supporting the clinical relevance of CIMP-positive tumors and suggest that a common CIMP etiology may define new mechanistic targets in cancer treatment.
Keywords: CpG island methylator phenotype; DNA methylation; pan-cancer.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Pan-cancer analysis reveals presence of pronounced DNA methylation drift in CpG island methylator phenotype clusters.Epigenomics. 2017 Nov;9(11):1341-1352. doi: 10.2217/epi-2017-0070. Epub 2017 Sep 29. Epigenomics. 2017. PMID: 28960094
-
Pan-cancer stratification of solid human epithelial tumors and cancer cell lines reveals commonalities and tissue-specific features of the CpG island methylator phenotype.Epigenetics Chromatin. 2015 Apr 17;8:14. doi: 10.1186/s13072-015-0007-7. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 25960768 Free PMC article.
-
Deciphering the etiology and role in oncogenic transformation of the CpG island methylator phenotype: a pan-cancer analysis.Brief Bioinform. 2022 Mar 10;23(2):bbab610. doi: 10.1093/bib/bbab610. Brief Bioinform. 2022. PMID: 35134107 Free PMC article.
-
Biological significance of the CpG island methylator phenotype.Biochem Biophys Res Commun. 2014 Dec 5;455(1-2):35-42. doi: 10.1016/j.bbrc.2014.07.007. Epub 2014 Jul 10. Biochem Biophys Res Commun. 2014. PMID: 25016183 Review.
-
Epigenetic alterations in colorectal cancer: the CpG island methylator phenotype.Histol Histopathol. 2013 May;28(5):585-95. doi: 10.14670/HH-28.585. Epub 2013 Jan 23. Histol Histopathol. 2013. PMID: 23341177 Review.
Cited by
-
Overview on Clinical Relevance of Intra-Tumor Heterogeneity.Front Med (Lausanne). 2018 Apr 6;5:85. doi: 10.3389/fmed.2018.00085. eCollection 2018. Front Med (Lausanne). 2018. PMID: 29682505 Free PMC article. Review.
-
Ferroptosis open a new door for colorectal cancer treatment.Front Oncol. 2023 Mar 16;13:1059520. doi: 10.3389/fonc.2023.1059520. eCollection 2023. Front Oncol. 2023. PMID: 37007121 Free PMC article. Review.
-
Targeted Assessment of G0S2 Methylation Identifies a Rapidly Recurrent, Routinely Fatal Molecular Subtype of Adrenocortical Carcinoma.Clin Cancer Res. 2019 Jun 1;25(11):3276-3288. doi: 10.1158/1078-0432.CCR-18-2693. Epub 2019 Feb 15. Clin Cancer Res. 2019. PMID: 30770352 Free PMC article.
-
Detecting Chromosome Instability in Cancer: Approaches to Resolve Cell-to-Cell Heterogeneity.Cancers (Basel). 2019 Feb 15;11(2):226. doi: 10.3390/cancers11020226. Cancers (Basel). 2019. PMID: 30781398 Free PMC article. Review.
-
Glioma CpG island methylator phenotype (G-CIMP): biological and clinical implications.Neuro Oncol. 2018 Apr 9;20(5):608-620. doi: 10.1093/neuonc/nox183. Neuro Oncol. 2018. PMID: 29036500 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

