Mechanisms for Complex Chromosomal Insertions
- PMID: 27880765
- PMCID: PMC5120786
- DOI: 10.1371/journal.pgen.1006446
Mechanisms for Complex Chromosomal Insertions
Abstract
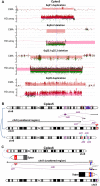
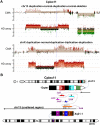
Chromosomal insertions are genomic rearrangements with a chromosome segment inserted into a non-homologous chromosome or a non-adjacent locus on the same chromosome or the other homologue, constituting ~2% of nonrecurrent copy-number gains. Little is known about the molecular mechanisms of their formation. We identified 16 individuals with complex insertions among 56,000 individuals tested at Baylor Genetics using clinical array comparative genomic hybridization (aCGH) and fluorescence in situ hybridization (FISH). Custom high-density aCGH was performed on 10 individuals with available DNA, and breakpoint junctions were fine-mapped at nucleotide resolution by long-range PCR and DNA sequencing in 6 individuals to glean insights into potential mechanisms of formation. We observed microhomologies and templated insertions at the breakpoint junctions, resembling the breakpoint junction signatures found in complex genomic rearrangements generated by replication-based mechanism(s) with iterative template switches. In addition, we analyzed 5 families with apparently balanced insertion in one parent detected by FISH analysis and found that 3 parents had additional small copy-number variants (CNVs) at one or both sides of the inserting fragments as well as at the inserted sites. We propose that replicative repair can result in interchromosomal complex insertions generated through chromothripsis-like chromoanasynthesis involving two or three chromosomes, and cause a significant fraction of apparently balanced insertions harboring small flanking CNVs.
Conflict of interest statement
JRL has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, Inc., is on the Scientific Advisory Board of Baylor Genetics, and is a co-inventor on multiple United States and European patents related to molecular diagnostics for inherited neuropathies, eye diseases and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Genetics (https://www.bcm.edu/research/medical-genetics-labs/).
Figures



References
-
- Van Hemel JO, Eussen HJ. Interchromosomal insertions. Identification of five cases and a review. Hum Genet. 2000;107(5):415–32. - PubMed
-
- Neill NJ, Ballif BC, Lamb AN, Parikh S, Ravnan JB, Schultz RA, et al. Recurrence, submicroscopic complexity, and potential clinical relevance of copy gains detected by array CGH that are shown to be unbalanced insertions by FISH. Genome Res. 2011;21(4):535–44. 10.1101/gr.114579.110 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

