Synonymous Co-Variation across the E1/E2 Gene Junction of Hepatitis C Virus Defines Virion Fitness
- PMID: 27880830
- PMCID: PMC5120871
- DOI: 10.1371/journal.pone.0167089
Synonymous Co-Variation across the E1/E2 Gene Junction of Hepatitis C Virus Defines Virion Fitness
Abstract
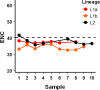
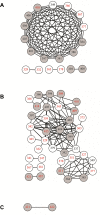
Hepatitis C virus is a positive-sense single-stranded RNA virus. The gene junction partitioning the viral glycoproteins E1 and E2 displays concurrent sequence evolution with the 3'-end of E1 highly conserved and the 5'-end of E2 highly heterogeneous. This gene junction is also believed to contain structured RNA elements, with a growing body of evidence suggesting that such structures can act as an additional level of viral replication and transcriptional control. We have previously used ultradeep pyrosequencing to analyze an amplicon library spanning the E1/E2 gene junction from a treatment naïve patient where samples were collected over 10 years of chronic HCV infection. During this timeframe maintenance of an in-frame insertion, recombination and humoral immune targeting of discrete virus sub-populations was reported. In the current study, we present evidence of epistatic evolution across the E1/E2 gene junction and observe the development of co-varying networks of codons set against a background of a complex virome with periodic shifts in population dominance. Overtime, the number of codons actively mutating decreases for all virus groupings. We identify strong synonymous co-variation between codon sites in a group of sequences harbouring a 3 bp in-frame insertion and propose that synonymous mutation acts to stabilize the RNA structural backbone.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures





Similar articles
-
Analysis of the evolution and structure of a complex intrahost viral population in chronic hepatitis C virus mapped by ultradeep pyrosequencing.J Virol. 2014 Dec;88(23):13709-21. doi: 10.1128/JVI.01732-14. Epub 2014 Sep 17. J Virol. 2014. PMID: 25231312 Free PMC article.
-
Hepatitis C Virus Envelope Glycoprotein E1 Forms Trimers at the Surface of the Virion.J Virol. 2015 Oct;89(20):10333-46. doi: 10.1128/JVI.00991-15. Epub 2015 Aug 5. J Virol. 2015. PMID: 26246575 Free PMC article.
-
Network Analysis of the Chronic Hepatitis C Virome Defines Hypervariable Region 1 Evolutionary Phenotypes in the Context of Humoral Immune Responses.J Virol. 2015 Dec 30;90(7):3318-29. doi: 10.1128/JVI.02995-15. J Virol. 2015. PMID: 26719263 Free PMC article.
-
Anti-envelope 1 and 2 immune response in chronic hepatitis C patients: effects of hepatitis B virus co-infection and interferon treatment.J Med Virol. 2004 May;73(1):33-7. doi: 10.1002/jmv.20058. J Med Virol. 2004. PMID: 15042645
-
[Tropism of hepatitis C virus for leukocytes-- importance of the analysis of viral E1 and E2 envelope glycoprotein genes by sequencing].Pathol Biol (Paris). 2010 Apr;58(2):170-4. doi: 10.1016/j.patbio.2009.06.010. Epub 2009 Nov 4. Pathol Biol (Paris). 2010. PMID: 19892492 Review. French.
References
-
- Steinhauer DA, Domingo E, Holland JJ. Lack of evidence for proofreading mechanisms associated with an RNA virus polymerase. Gene. 1992;122(2):281–8. . - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

