Highly Divergent Clostridium difficile Strains Isolated from the Environment
- PMID: 27880843
- PMCID: PMC5120845
- DOI: 10.1371/journal.pone.0167101
Highly Divergent Clostridium difficile Strains Isolated from the Environment
Abstract
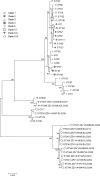
Clostridium difficile is one of the most important human and animal pathogens. However, the bacterium is ubiquitous and can be isolated from various sources. Here we report the prevalence and characterization of C. difficile in less studied environmental samples, puddle water (n = 104) and soil (n = 79). C. difficile was detected in 14.4% of puddle water and in 36.7% of soil samples. Environmental strains displayed antimicrobial resistance patterns comparable to already published data of human and animal isolates. A total of 480 isolates were grouped into 34 different PCR ribotypes. More than half of these (52.9%; 18 of 34) were already described in humans or animals. However, 14 PCR ribotypes were new in our PCR ribotype library and all but one were non-toxigenic. The multilocus sequence analysis of these new PCR ribotypes revealed that non-toxigenic environmental isolates are phylogenetically distinct and belong to three highly divergent clades, two of which have not been described before. Our data suggest that environment is a potential reservoir of genetically diverse population of C. difficile.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

