Formation and processing of DNA damage substrates for the hNEIL enzymes
- PMID: 27880870
- PMCID: PMC5438787
- DOI: 10.1016/j.freeradbiomed.2016.11.030
Formation and processing of DNA damage substrates for the hNEIL enzymes
Abstract
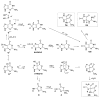
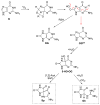
Reactive oxygen species (ROS) are harnessed by the cell for signaling at the same time as being detrimental to cellular components such as DNA. The genome and transcriptome contain instructions that can alter cellular processes when oxidized. The guanine (G) heterocycle in the nucleotide pool, DNA, or RNA is the base most prone to oxidation. The oxidatively-derived products of G consistently observed in high yields from hydroxyl radical, carbonate radical, or singlet oxygen oxidations under conditions modeling the cellular reducing environment are discussed. The major G base oxidation products are 8-oxo-7,8-dihydroguanine (OG), 5-carboxamido-5-formamido-2-iminohydantoin (2Ih), spiroiminodihydantoin (Sp), and 5-guanidinohydantoin (Gh). The yields of these products show dependency on the oxidant and the reaction context that includes nucleoside, single-stranded DNA (ssDNA), double-stranded DNA (dsDNA), and G-quadruplex DNA (G4-DNA) structures. Upon formation of these products in cells, they are recognized by the DNA glycosylases in the base excision repair (BER) pathway. This review focuses on initiation of BER by the mammalian Nei-like1-3 (NEIL1-3) glycosylases for removal of 2Ih, Sp, and Gh. The unique ability of the human NEILs to initiate removal of the hydantoins in ssDNA, bulge-DNA, bubble-DNA, dsDNA, and G4-DNA is outlined. Additionally, when Gh exists in a G4 DNA found in a gene promoter, NEIL-mediated repair is modulated by the plasticity of the G4-DNA structure provided by additional G-runs flanking the sequence. On the basis of these observations and cellular studies from the literature, the interplay between DNA oxidation and BER to alter gene expression is discussed.
Keywords: 5-carboxamido-5-formamido-2-iminohydantoin; 5-guanidinohydantoin; 8-oxo-7,8-dihydroguanine; G-quadruplex; NEIL DNA glycosylase; Spiroiminodihydantoin.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Removal of oxidatively generated DNA damage by overlapping repair pathways.Free Radic Biol Med. 2017 Jun;107:53-61. doi: 10.1016/j.freeradbiomed.2016.10.507. Epub 2016 Nov 4. Free Radic Biol Med. 2017. PMID: 27818219 Free PMC article. Review.
-
Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2.DNA Repair (Amst). 2005 Jan 2;4(1):41-50. doi: 10.1016/j.dnarep.2004.07.006. DNA Repair (Amst). 2005. PMID: 15533836
-
G-quadruplex folds of the human telomere sequence alter the site reactivity and reaction pathway of guanine oxidation compared to duplex DNA.Chem Res Toxicol. 2013 Apr 15;26(4):593-607. doi: 10.1021/tx400028y. Epub 2013 Mar 13. Chem Res Toxicol. 2013. PMID: 23438298 Free PMC article.
-
2'-Fluorinated Hydantoins as Chemical Biology Tools for Base Excision Repair Glycosylases.ACS Chem Biol. 2020 Apr 17;15(4):915-924. doi: 10.1021/acschembio.9b00923. Epub 2020 Mar 13. ACS Chem Biol. 2020. PMID: 32069022 Free PMC article.
-
Oxidative DNA damage repair in mammalian cells: a new perspective.DNA Repair (Amst). 2007 Apr 1;6(4):470-80. doi: 10.1016/j.dnarep.2006.10.011. Epub 2006 Nov 20. DNA Repair (Amst). 2007. PMID: 17116430 Free PMC article. Review.
Cited by
-
Case studies on potential G-quadruplex-forming sequences from the bacterial orders Deinococcales and Thermales derived from a survey of published genomes.Sci Rep. 2018 Oct 24;8(1):15679. doi: 10.1038/s41598-018-33944-4. Sci Rep. 2018. PMID: 30356061 Free PMC article.
-
The RAD17 Promoter Sequence Contains a Potential Tail-Dependent G-Quadruplex That Downregulates Gene Expression upon Oxidative Modification.ACS Chem Biol. 2018 Sep 21;13(9):2577-2584. doi: 10.1021/acschembio.8b00522. Epub 2018 Aug 15. ACS Chem Biol. 2018. PMID: 30063821 Free PMC article.
-
Formation and repair of oxidatively generated damage in cellular DNA.Free Radic Biol Med. 2017 Jun;107:13-34. doi: 10.1016/j.freeradbiomed.2016.12.049. Epub 2017 Jan 2. Free Radic Biol Med. 2017. PMID: 28057600 Free PMC article. Review.
-
Unhooking of an interstrand cross-link at DNA fork structures by the DNA glycosylase NEIL3.DNA Repair (Amst). 2020 Feb;86:102752. doi: 10.1016/j.dnarep.2019.102752. Epub 2019 Nov 20. DNA Repair (Amst). 2020. PMID: 31923807 Free PMC article.
-
The Influence of Oxidized Imino-Allantoin in the Presence of OXOG on Double Helix Charge Transfer: A Theoretical Approach.Int J Mol Sci. 2024 May 29;25(11):5962. doi: 10.3390/ijms25115962. Int J Mol Sci. 2024. PMID: 38892152 Free PMC article.
References
-
- Giorgio M, Trinei M, Migliaccio E, Pelicci PG. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat Rev Mol Cell Biol. 2007;8:722–728. - PubMed
-
- Speckmann B, Steinbrenner H, Grune T, Klotz LO. Peroxynitrite: From interception to signaling. Arch Biochem Biophys. 2016;595:153–160. - PubMed
-
- Dedon PC, Tannenbaum SR. Reactive nitrogen species in the chemical biology of inflammation. Arch Biochem Biophys. 2004;423:12–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

