LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage
- PMID: 27880904
- PMCID: PMC6940422
- DOI: 10.1016/j.celrep.2016.10.080
LYVE1 Marks the Divergence of Yolk Sac Definitive Hemogenic Endothelium from the Primitive Erythroid Lineage
Abstract
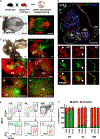
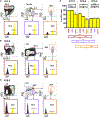
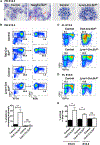
The contribution of the different waves and sites of developmental hematopoiesis to fetal and adult blood production remains unclear. Here, we identify lymphatic vessel endothelial hyaluronan receptor-1 (LYVE1) as a marker of yolk sac (YS) endothelium and definitive hematopoietic stem and progenitor cells (HSPCs). Endothelium in mid-gestation YS and vitelline vessels, but not the dorsal aorta and placenta, were labeled by Lyve1-Cre. Most YS HSPCs and erythro-myeloid progenitors were Lyve1-Cre lineage traced, but primitive erythroid cells were not, suggesting that they represent distinct lineages. Fetal liver (FL) and adult HSPCs showed 35%-40% Lyve1-Cre marking. Analysis of circulation-deficient Ncx1-/- concepti identified the YS as a major source of Lyve1-Cre labeled HSPCs. FL proerythroblast marking was extensive at embryonic day (E) 11.5-13.5, but decreased to hematopoietic stem cell (HSC) levels by E16.5, suggesting that HSCs from multiple sources became responsible for erythropoiesis. Lyve1-Cre thus marks the divergence between YS primitive and definitive hematopoiesis and provides a tool for targeting YS definitive hematopoiesis and FL colonization.
Keywords: LYVE1; definitive hematopoiesis; fetal liver; hemogenic endothelium; lineage tracing; primitive hematopoiesis; yolk sac.
Copyright © 2016 The Authors. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Ezh2 is essential for the generation of functional yolk sac derived erythro-myeloid progenitors.Nat Commun. 2021 Dec 2;12(1):7019. doi: 10.1038/s41467-021-27140-8. Nat Commun. 2021. PMID: 34857757 Free PMC article.
-
Mds1CreERT2, an inducible Cre allele specific to adult-repopulating hematopoietic stem cells.Cell Rep. 2021 Aug 17;36(7):109562. doi: 10.1016/j.celrep.2021.109562. Cell Rep. 2021. PMID: 34407416 Free PMC article.
-
Notch1 regulates progenitor cell proliferation and differentiation during mouse yolk sac hematopoiesis.Cell Death Differ. 2014 Jul;21(7):1081-94. doi: 10.1038/cdd.2014.27. Epub 2014 Feb 28. Cell Death Differ. 2014. PMID: 24583642 Free PMC article.
-
Hematopoiesis in the yolk sac: more than meets the eye.Exp Hematol. 2005 Sep;33(9):1021-8. doi: 10.1016/j.exphem.2005.06.012. Exp Hematol. 2005. PMID: 16140150 Review.
-
Cellular Basis of Embryonic Hematopoiesis and Its Implications in Prenatal Erythropoiesis.Int J Mol Sci. 2020 Dec 8;21(24):9346. doi: 10.3390/ijms21249346. Int J Mol Sci. 2020. PMID: 33302450 Free PMC article. Review.
Cited by
-
Hyaluronic Acid: Known for Almost a Century, but Still in Vogue.Pharmaceutics. 2022 Apr 11;14(4):838. doi: 10.3390/pharmaceutics14040838. Pharmaceutics. 2022. PMID: 35456670 Free PMC article. Review.
-
Stem Cell-Based Disease Models for Inborn Errors of Immunity.Cells. 2021 Dec 30;11(1):108. doi: 10.3390/cells11010108. Cells. 2021. PMID: 35011669 Free PMC article. Review.
-
Single-Cell RNA Sequencing Delineates the Atlas and Cell Interactions of the Testicular Cells in Chinese Soft-Shelled Turtle (Pelodiscus sinensis).Mar Biotechnol (NY). 2025 Jun 23;27(4):101. doi: 10.1007/s10126-025-10474-5. Mar Biotechnol (NY). 2025. PMID: 40549204
-
Murine hematopoietic stem cell activity is derived from pre-circulation embryos but not yolk sacs.Nat Commun. 2018 Dec 20;9(1):5405. doi: 10.1038/s41467-018-07769-8. Nat Commun. 2018. PMID: 30573729 Free PMC article.
-
Hyperactive KRAS/MAPK signaling disrupts normal lymphatic vessel architecture and function.Front Cell Dev Biol. 2023 Sep 25;11:1276333. doi: 10.3389/fcell.2023.1276333. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37842094 Free PMC article.
References
-
- Benz C, Copley MR, Kent DG, Wohrer S, Cortes A, Aghaeepour N, Ma E, Mader H, Rowe K, Day C, et al. (2012). Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell 10, 273–283. - PubMed
-
- Boisset JC, van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, and Robin C (2010). In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 464, 116–120. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

