Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores
- PMID: 27881301
- PMCID: PMC5101189
- DOI: 10.1016/j.cell.2016.10.005
Structure of the MIS12 Complex and Molecular Basis of Its Interaction with CENP-C at Human Kinetochores
Abstract
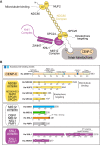
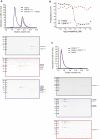
Kinetochores, multisubunit protein assemblies, connect chromosomes to spindle microtubules to promote chromosome segregation. The 10-subunit KMN assembly (comprising KNL1, MIS12, and NDC80 complexes, designated KNL1C, MIS12C, and NDC80C) binds microtubules and regulates mitotic checkpoint function through NDC80C and KNL1C, respectively. MIS12C, on the other hand, connects the KMN to the chromosome-proximal domain of the kinetochore through a direct interaction with CENP-C. The structural basis for this crucial bridging function of MIS12C is unknown. Here, we report crystal structures of human MIS12C associated with a fragment of CENP-C and unveil the role of Aurora B kinase in the regulation of this interaction. The structure of MIS12:CENP-C complements previously determined high-resolution structures of functional regions of NDC80C and KNL1C and allows us to build a near-complete structural model of the KMN assembly. Our work illuminates the structural organization of essential chromosome segregation machinery that is conserved in most eukaryotes.
Keywords: CCAN; CENP-C; DSN1; KMN network; MIND; Mis12; NSL1; PMF1; centromere; kinetochore.
Copyright © 2016 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures











Comment in
-
MIS12/MIND Control at the Kinetochore.Cell. 2016 Nov 3;167(4):889-891. doi: 10.1016/j.cell.2016.10.036. Cell. 2016. PMID: 27814517
Similar articles
-
Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores.J Cell Biol. 2015 Jan 19;208(2):181-96. doi: 10.1083/jcb.201407074. J Cell Biol. 2015. PMID: 25601404 Free PMC article.
-
Structure of the human outer kinetochore KMN network complex.Nat Struct Mol Biol. 2024 Jun;31(6):874-883. doi: 10.1038/s41594-024-01249-y. Epub 2024 Mar 8. Nat Struct Mol Biol. 2024. PMID: 38459127 Free PMC article.
-
Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T.Curr Biol. 2015 Mar 2;25(5):671-7. doi: 10.1016/j.cub.2015.01.059. Epub 2015 Feb 5. Curr Biol. 2015. PMID: 25660545 Free PMC article.
-
The kinetochore interaction network (KIN) of ascomycetes.Mycologia. 2016 May-Jun;108(3):485-505. doi: 10.3852/15-182. Epub 2016 Feb 23. Mycologia. 2016. PMID: 26908646 Free PMC article. Review.
-
Where is the right path heading from the centromere to spindle microtubules?Cell Cycle. 2019 Jun;18(11):1199-1211. doi: 10.1080/15384101.2019.1617008. Epub 2019 May 20. Cell Cycle. 2019. PMID: 31075048 Free PMC article. Review.
Cited by
-
Dephosphorylation in nuclear reassembly after mitosis.Front Cell Dev Biol. 2022 Oct 4;10:1012768. doi: 10.3389/fcell.2022.1012768. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36268509 Free PMC article. Review.
-
On the Regulation of Mitosis by the Kinetochore, a Macromolecular Complex and Organising Hub of Eukaryotic Organisms.Subcell Biochem. 2022;99:235-267. doi: 10.1007/978-3-031-00793-4_7. Subcell Biochem. 2022. PMID: 36151378
-
CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization.J Cell Biol. 2019 Dec 2;218(12):4042-4062. doi: 10.1083/jcb.201907006. Epub 2019 Nov 1. J Cell Biol. 2019. PMID: 31676716 Free PMC article.
-
CDC20 in and out of mitosis: a prognostic factor and therapeutic target in hematological malignancies.J Exp Clin Cancer Res. 2022 Apr 30;41(1):159. doi: 10.1186/s13046-022-02363-9. J Exp Clin Cancer Res. 2022. PMID: 35490245 Free PMC article. Review.
-
Tight bending of the Ndc80 complex provides intrinsic regulation of its binding to microtubules.Elife. 2019 May 2;8:e44489. doi: 10.7554/eLife.44489. Elife. 2019. PMID: 31045495 Free PMC article.
References
-
- Barth T.K., Schade G.O., Schmidt A., Vetter I., Wirth M., Heun P., Thomae A.W., Imhof A. Identification of novel Drosophila centromere-associated proteins. Proteomics. 2014;14:2167–2178. - PubMed
-
- Bharadwaj R., Qi W., Yu H. Identification of two novel components of the human NDC80 kinetochore complex. J. Biol. Chem. 2004;279:13076–13085. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

