Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket
- PMID: 27881302
- PMCID: PMC5127602
- DOI: 10.1016/j.cell.2016.09.056
Crystal Structure of a Full-Length Human Tetraspanin Reveals a Cholesterol-Binding Pocket
Abstract
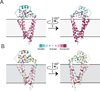
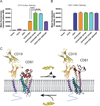
Tetraspanins comprise a diverse family of four-pass transmembrane proteins that play critical roles in the immune, reproductive, genitourinary, and auditory systems. Despite their pervasive roles in human physiology, little is known about the structure of tetraspanins or the molecular mechanisms underlying their various functions. Here, we report the crystal structure of human CD81, a full-length tetraspanin. The transmembrane segments of CD81 pack as two largely separated pairs of helices, capped by the large extracellular loop (EC2) at the outer membrane leaflet. The two pairs of helices converge at the inner leaflet to create an intramembrane pocket with additional electron density corresponding to a bound cholesterol molecule within the cavity. Molecular dynamics simulations identify an additional conformation in which EC2 separates substantially from the transmembrane domain. Cholesterol binding appears to modulate CD81 activity in cells, suggesting a potential mechanism for regulation of tetraspanin function.
Keywords: CD19; CD81; X-ray crystallography; membrane protein; protein structure; protein trafficking.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures



References
-
- Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. Journal of chemical theory and computation. 2012b;8:3257–3273. - PMC - PubMed
-
- Bradbury LE, Kansas GS, Levy S, Evans RL, Tedder TF. The CD19/CD21 signal transducing complex of human B lymphocytes includes the target of antiproliferative antibody-1 and Leu-13 molecules. Journal of immunology. 1992;149:2841–2850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

