Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences
- PMID: 27881428
- PMCID: PMC6018996
- DOI: 10.1093/bib/bbw114
Genome, transcriptome and proteome: the rise of omics data and their integration in biomedical sciences
Abstract
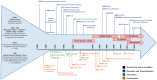
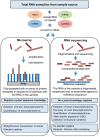
Advances in the technologies and informatics used to generate and process large biological data sets (omics data) are promoting a critical shift in the study of biomedical sciences. While genomics, transcriptomics and proteinomics, coupled with bioinformatics and biostatistics, are gaining momentum, they are still, for the most part, assessed individually with distinct approaches generating monothematic rather than integrated knowledge. As other areas of biomedical sciences, including metabolomics, epigenomics and pharmacogenomics, are moving towards the omics scale, we are witnessing the rise of inter-disciplinary data integration strategies to support a better understanding of biological systems and eventually the development of successful precision medicine. This review cuts across the boundaries between genomics, transcriptomics and proteomics, summarizing how omics data are generated, analysed and shared, and provides an overview of the current strengths and weaknesses of this global approach. This work intends to target students and researchers seeking knowledge outside of their field of expertise and fosters a leap from the reductionist to the global-integrative analytical approach in research.
Figures




References
-
- Bernfield MR, Nirenberg MW.. RNA codewords and protein synthesis. the nucleotide sequences of multiple codewords for phenylalanine, serine, leucine, and proline. Science 1965;147:479–84. - PubMed
-
- Genomes. http://www.1000genomes.org/.
-
- International HapMap Consortium. The International HapMap project. Nature 2003;426:789–96. - PubMed
-
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature 2004;431:931–45. - PubMed
-
- Protein D. The First Solution of the Three-Dimensional Molecular Structure of a Protein (1958 – 1960). HistoryofInformation.com.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

