Predictive approaches for drug combination discovery in cancer
- PMID: 27881431
- PMCID: PMC6018991
- DOI: 10.1093/bib/bbw104
Predictive approaches for drug combination discovery in cancer
Abstract
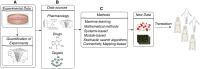
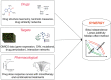
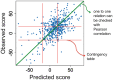
Drug combinations have been proposed as a promising therapeutic strategy to overcome drug resistance and improve efficacy of monotherapy regimens in cancer. This strategy aims at targeting multiple components of this complex disease. Despite the increasing number of drug combinations in use, many of them were empirically found in the clinic, and the molecular mechanisms underlying these drug combinations are often unclear. These challenges call for rational, systematic approaches for drug combination discovery. Although high-throughput screening of single-agent therapeutics has been successfully implemented, it is not feasible to test all possible drug combinations, even for a reduced subset of anticancer drugs. Hence, in vitro and in vivo screening of a large number of drug combinations are not practical. Therefore, devising computational methods to efficiently explore the space of drug combinations and to discover efficacious combinations has attracted a lot of attention from the scientific community in the past few years. Nevertheless, in the absence of consensus regarding the computational approaches used to predict efficacious drug combinations, a plethora of methods, techniques and hypotheses have been developed to date, while the research field lacks an elaborate categorization of the existing computational methods and the available data sources. In this manuscript, we review and categorize the state-of-the-art computational approaches for drug combination prediction, and elaborate on the limitations of these methods and the existing challenges. We also discuss about the recent pan-cancer drug combination data sets and their importance in revising the available methods or developing more performant approaches.
Figures

References
-
- Zimmermann GR, Lehár J, Keith CT.. Multi-target therapeutics: when the whole is greater than the sum of the parts. Drug Discov Today 2007;12:34–42. - PubMed
-
- Gottesman MM. Mechanisms of cancer drug resistance. Annu Rev Med 2002;53:615–27. - PubMed
-
- DeVita VT Jr, Young RC, Canellos GP.. Combination versus single agent chemotherapy: a review of the basis for selection of drug treatment of cancer. Cancer 1975;35:98–110. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

