Casein kinase II is required for proper cell division and acts as a negative regulator of centrosome duplication in Caenorhabditis elegans embryos
- PMID: 27881437
- PMCID: PMC5278433
- DOI: 10.1242/bio.022418
Casein kinase II is required for proper cell division and acts as a negative regulator of centrosome duplication in Caenorhabditis elegans embryos
Abstract
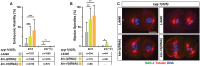
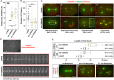
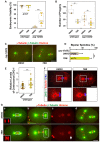
Centrosomes are the primary microtubule-organizing centers that orchestrate microtubule dynamics during the cell cycle. The correct number of centrosomes is pivotal for establishing bipolar mitotic spindles that ensure accurate segregation of chromosomes. Thus, centrioles must duplicate once per cell cycle, one daughter per mother centriole, the process of which requires highly coordinated actions among core factors and modulators. Protein phosphorylation is shown to regulate the stability, localization and activity of centrosome proteins. Here, we report the function of Casein kinase II (CK2) in early Caenorhabditis elegans embryos. The catalytic subunit (KIN-3/CK2α) of CK2 localizes to nuclei, centrosomes and midbodies. Inactivating CK2 leads to cell division defects, including chromosome missegregation, cytokinesis failure and aberrant centrosome behavior. Furthermore, depletion or inhibiting kinase activity of CK2 results in elevated ZYG-1 levels at centrosomes, restoring centrosome duplication and embryonic viability to zyg-1 mutants. Our data suggest that CK2 functions in cell division and negatively regulates centrosome duplication in a kinase-dependent manner.
Keywords: CK2; Caenorhabditis elegans; Casein kinase II; Centrosome; KIN-3; ZYG-1.
© 2017. Published by The Company of Biologists Ltd.
Conflict of interest statement
The authors declare no competing or financial interests.
Figures



Similar articles
-
Site-specific phosphorylation of ZYG-1 regulates ZYG-1 stability and centrosome number.iScience. 2023 Nov 8;26(12):108410. doi: 10.1016/j.isci.2023.108410. eCollection 2023 Dec 15. iScience. 2023. PMID: 38034351 Free PMC article.
-
APC/CFZR-1 Controls SAS-5 Levels To Regulate Centrosome Duplication in Caenorhabditis elegans.G3 (Bethesda). 2017 Dec 4;7(12):3937-3946. doi: 10.1534/g3.117.300260. G3 (Bethesda). 2017. PMID: 29030390 Free PMC article.
-
The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo.Cell. 2001 May 18;105(4):547-58. doi: 10.1016/s0092-8674(01)00338-5. Cell. 2001. PMID: 11371350
-
Centrosome duplication: of rules and licenses.Trends Cell Biol. 2007 May;17(5):215-21. doi: 10.1016/j.tcb.2007.03.003. Epub 2007 Mar 26. Trends Cell Biol. 2007. PMID: 17383880 Review.
-
Centrosome maturation - in tune with the cell cycle.J Cell Sci. 2022 Jan 15;135(2):jcs259395. doi: 10.1242/jcs.259395. Epub 2022 Jan 28. J Cell Sci. 2022. PMID: 35088834 Review.
Cited by
-
Minor Kinases with Major Roles in Cytokinesis Regulation.Cells. 2022 Nov 17;11(22):3639. doi: 10.3390/cells11223639. Cells. 2022. PMID: 36429067 Free PMC article. Review.
-
The C. elegans Casein Kinase II is associated with meiotic DNA in fertilized oocytes.MicroPubl Biol. 2022 Jun 6;2022:10.17912/micropub.biology.000583. doi: 10.17912/micropub.biology.000583. eCollection 2022. MicroPubl Biol. 2022. PMID: 35685274 Free PMC article.
-
Site-specific phosphorylation of ZYG-1 regulates ZYG-1 stability and centrosome number.iScience. 2023 Nov 8;26(12):108410. doi: 10.1016/j.isci.2023.108410. eCollection 2023 Dec 15. iScience. 2023. PMID: 38034351 Free PMC article.
-
Conservation of OFD1 Protein Motifs: Implications for Discovery of Novel Interactors and the OFD1 Function.Int J Mol Sci. 2025 Jan 29;26(3):1167. doi: 10.3390/ijms26031167. Int J Mol Sci. 2025. PMID: 39940934 Free PMC article.
-
APC/CFZR-1 regulates centrosomal ZYG-1 to limit centrosome number.J Cell Sci. 2021 Jul 15;134(14):jcs253088. doi: 10.1242/jcs.253088. Epub 2021 Jul 26. J Cell Sci. 2021. PMID: 34308970 Free PMC article.
References
-
- Alessi A. F., Khivansara V., Han T., Freeberg M. A., Moresco J. J., Tu P. G., Montoye E., Yates J. R. III, Karp X. and Kim J. K. (2015). Casein kinase II promotes target silencing by miRISC through direct phosphorylation of the DEAD-box RNA helicase CGH-1. Proc. Natl. Acad. Sci. USA 112, E7213-E7222. 10.1073/pnas.1509499112 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

