Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G
- PMID: 27881650
- PMCID: PMC5244329
- DOI: 10.1128/JVI.02230-16
Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G
Erratum in
-
Erratum for Ara et al., "Mechanism of Enhanced HIV Restriction by Virion Coencapsidated Cytidine Deaminases APOBEC3F and APOBEC3G".J Virol. 2025 Feb 25;99(2):e0220424. doi: 10.1128/jvi.02204-24. Epub 2025 Jan 16. J Virol. 2025. PMID: 39817775 Free PMC article. No abstract available.
Abstract
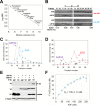
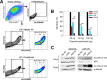
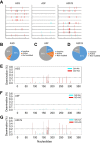
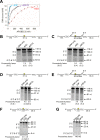
The APOBEC3 (A3) enzymes, A3G and A3F, are coordinately expressed in CD4+ T cells and can become coencapsidated into HIV-1 virions, primarily in the absence of the viral infectivity factor (Vif). A3F and A3G are deoxycytidine deaminases that inhibit HIV-1 replication by inducing guanine-to-adenine hypermutation through deamination of cytosine to form uracil in minus-strand DNA. The effect of the simultaneous presence of both A3G and A3F on HIV-1 restriction ability is not clear. Here, we used a single-cycle infectivity assay and biochemical analyses to determine if coencapsidated A3G and A3F differ in their restriction capacity from A3G or A3F alone. Proviral DNA sequencing demonstrated that compared to each A3 enzyme alone, A3G and A3F, when combined, had a coordinate effect on hypermutation. Using size exclusion chromatography, rotational anisotropy, and in vitro deamination assays, we demonstrate that A3F promotes A3G deamination activity by forming an A3F/G hetero-oligomer in the absence of RNA which is more efficient at deaminating cytosines. Further, A3F caused the accumulation of shorter reverse transcripts due to decreasing reverse transcriptase efficiency, which would leave single-stranded minus-strand DNA exposed for longer periods of time, enabling more deamination events to occur. Although A3G and A3F are known to function alongside each other, these data provide evidence for an A3F/G hetero-oligomeric A3 with unique properties compared to each individual counterpart.
Importance: The APOBEC3 enzymes APOBEC3F and APOBEC3G act as a barrier to HIV-1 replication in the absence of the HIV-1 Vif protein. After APOBEC3 enzymes are encapsidated into virions, they deaminate cytosines in minus-strand DNA, which forms promutagenic uracils that induce transition mutations or proviral DNA degradation. Even in the presence of Vif, footprints of APOBEC3-catalyzed deaminations are found, demonstrating that APOBEC3s still have discernible activity against HIV-1 in infected individuals. We undertook a study to better understand the activity of coexpressed APOBEC3F and APOBEC3G. The data demonstrate that an APOBEC3F/APOBEC3G hetero-oligomer can form that has unique properties compared to each APOBEC3 alone. This hetero-oligomer has increased efficiency of virus hypermutation, raising the idea that we still may not fully realize the antiviral mechanisms of endogenous APOBEC3 enzymes. Hetero-oligomerization may be a mechanism to increase their antiviral activity in the presence of Vif.
Keywords: APOBEC3; DNA-protein interactions; HIV; deaminase; mutagenesis; oligomerization; processivity; protein-protein interactions; reverse transcriptase.
Copyright © 2017 American Society for Microbiology.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

