Human selenoprotein P and S variant mRNAs with different numbers of SECIS elements and inferences from mutant mice of the roles of multiple SECIS elements
- PMID: 27881738
- PMCID: PMC5133445
- DOI: 10.1098/rsob.160241
Human selenoprotein P and S variant mRNAs with different numbers of SECIS elements and inferences from mutant mice of the roles of multiple SECIS elements
Abstract
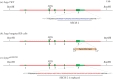
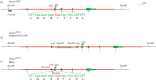
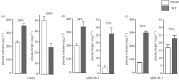
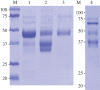
Dynamic redefinition of the 10 UGAs in human and mouse selenoprotein P (Sepp1) mRNAs to specify selenocysteine instead of termination involves two 3' UTR structural elements (SECIS) and is regulated by selenium availability. In addition to the previously known human Sepp1 mRNA poly(A) addition site just 3' of SECIS 2, two further sites were identified with one resulting in 10-25% of the mRNA lacking SECIS 2. To address function, mutant mice were generated with either SECIS 1 or SECIS 2 deleted or with the first UGA substituted with a serine codon. They were fed on either high or selenium-deficient diets. The mutants had very different effects on the proportions of shorter and longer product Sepp1 protein isoforms isolated from plasma, and on viability. Spatially and functionally distinctive effects of the two SECIS elements on UGA decoding were inferred. We also bioinformatically identify two selenoprotein S mRNAs with different 5' sequences predicted to yield products with different N-termini. These results provide insights into SECIS function and mRNA processing in selenoprotein isoform diversity.
Keywords: codon redefinition; ribosome specialization; selenocysteine; selenoprotein P; selenoprotein S.
© 2016 The Authors.
Figures



Similar articles
-
Efficient incorporation of multiple selenocysteines involves an inefficient decoding step serving as a potential translational checkpoint and ribosome bottleneck.Mol Cell Biol. 2006 Dec;26(24):9177-84. doi: 10.1128/MCB.00856-06. Epub 2006 Sep 25. Mol Cell Biol. 2006. PMID: 17000762 Free PMC article.
-
Regulation of selenocysteine incorporation into the selenium transport protein, selenoprotein P.J Biol Chem. 2014 Sep 5;289(36):25317-26. doi: 10.1074/jbc.M114.590430. Epub 2014 Jul 25. J Biol Chem. 2014. PMID: 25063811 Free PMC article.
-
Multiple RNA structures affect translation initiation and UGA redefinition efficiency during synthesis of selenoprotein P.Nucleic Acids Res. 2017 Dec 15;45(22):13004-13015. doi: 10.1093/nar/gkx982. Nucleic Acids Res. 2017. PMID: 29069514 Free PMC article.
-
The Interaction between Dietary Selenium Intake and Genetics in Determining Cancer Risk and Outcome.Nutrients. 2020 Aug 12;12(8):2424. doi: 10.3390/nu12082424. Nutrients. 2020. PMID: 32806741 Free PMC article. Review.
-
Solution structure of SECIS, the mRNA element required for eukaryotic selenocysteine insertion--interaction studies with the SECIS-binding protein SBP.Biomed Environ Sci. 1997 Sep;10(2-3):177-81. Biomed Environ Sci. 1997. PMID: 9315308 Review.
Cited by
-
RefSeq curation and annotation of stop codon recoding in vertebrates.Nucleic Acids Res. 2019 Jan 25;47(2):594-606. doi: 10.1093/nar/gky1234. Nucleic Acids Res. 2019. PMID: 30535227 Free PMC article.
-
Selenoproteins and the senescence-associated epitranscriptome.Exp Biol Med (Maywood). 2022 Dec;247(23):2090-2102. doi: 10.1177/15353702221116592. Epub 2022 Aug 29. Exp Biol Med (Maywood). 2022. PMID: 36036467 Free PMC article. Review.
-
New Directions for Understanding the Codon Redefinition Required for Selenocysteine Incorporation.Biol Trace Elem Res. 2019 Nov;192(1):18-25. doi: 10.1007/s12011-019-01827-y. Epub 2019 Jul 24. Biol Trace Elem Res. 2019. PMID: 31342342 Free PMC article. Review.
-
From Recoding to Peptides for MHC Class I Immune Display: Enriching Viral Expression, Virus Vulnerability and Virus Evasion.Viruses. 2021 Jun 27;13(7):1251. doi: 10.3390/v13071251. Viruses. 2021. PMID: 34199077 Free PMC article. Review.
-
Processive Recoding and Metazoan Evolution of Selenoprotein P: Up to 132 UGAs in Molluscs.J Mol Biol. 2019 Nov 8;431(22):4381-4407. doi: 10.1016/j.jmb.2019.08.007. Epub 2019 Aug 20. J Mol Biol. 2019. PMID: 31442478 Free PMC article.
References
-
- Mukai T, Englert M, Tripp HJ, Miller C, Ivanova NN, Rubin EM, Kyrpides NC, Söll D. 2016. Facile recoding of selenocysteine in nature. Angew. Chem. Int. Ed. Engl. 55, 5337–5341. (doi:10.1002/anie.201511657) - DOI - PMC - PubMed
-
- Hill KE, Lloyd RS, Yang JG, Read R, Burk RF. 1991. The cDNA for rat selenoprotein P contains 10 TGA codons in the open reading frame. J. Biol. Chem. 266, 10 050–10 053. - PubMed
-
- Hill KE, Lloyd RS, Burk RF. 1993. Conserved nucleotide sequences in the open reading frame and 3' untranslated region of selenoprotein P mRNA. Proc. Natl Acad. Sci. USA 90, 537–541. (doi:10.1073/pnas.90.2.537) - DOI - PMC - PubMed
-
- Kollmus H, Flohé L, McCarthy JE. 1996. Analysis of eukaryotic mRNA structures directing cotranslational incorporation of selenocysteine. Nucleic Acids Res. 24, 1195–1201. (doi:10.1093/nar/24.7.1195) - DOI - PMC - PubMed
-
- Gesteland RF, Atkins JF. 1996. Recoding: dynamic reprogramming of translation. Annu. Rev. Biochem. 65, 741–768. (doi:10.1146/annurev.bi.65.070196.003521) - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

