The Oxidosqualene Cyclase from the Oomycete Saprolegnia parasitica Synthesizes Lanosterol as a Single Product
- PMID: 27881978
- PMCID: PMC5101207
- DOI: 10.3389/fmicb.2016.01802
The Oxidosqualene Cyclase from the Oomycete Saprolegnia parasitica Synthesizes Lanosterol as a Single Product
Abstract
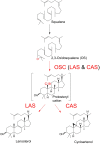
The first committed step of sterol biosynthesis is the cyclisation of 2,3-oxidosqualene to form either lanosterol (LA) or cycloartenol (CA). This is catalyzed by an oxidosqualene cyclase (OSC). LA and CA are subsequently converted into various sterols by a series of enzyme reactions. The specificity of the OSC therefore determines the final composition of the end sterols of an organism. Despite the functional importance of OSCs, the determinants of their specificity are not well understood. In sterol-synthesizing oomycetes, recent bioinformatics, and metabolite analysis suggest that LA is produced. However, this catalytic activity has never been experimentally demonstrated. Here, we show that the OSC of the oomycete Saprolegnia parasitica, a severe pathogen of salmonid fish, has an uncommon sequence in a conserved motif important for specificity. We present phylogenetic analysis revealing that this sequence is common to sterol-synthesizing oomycetes, as well as some plants, and hypothesize as to the evolutionary origin of some microbial sequences. We also demonstrate for the first time that a recombinant form of the OSC from S. parasitica produces LA exclusively. Our data pave the way for a detailed structural characterization of the protein and the possible development of specific inhibitors of oomycete OSCs for disease control in aquaculture.
Keywords: Saprolegnia parasitica; lanosterol biosynthesis; oomycete; oxidosqualene cyclase; sterols.
Figures




Similar articles
-
Comparative analysis of sterol acquisition in the oomycetes Saprolegnia parasitica and Phytophthora infestans.PLoS One. 2017 Feb 2;12(2):e0170873. doi: 10.1371/journal.pone.0170873. eCollection 2017. PLoS One. 2017. PMID: 28152045 Free PMC article.
-
Expressed sequence tags from the oomycete fish pathogen Saprolegnia parasitica reveal putative virulence factors.BMC Microbiol. 2005 Aug 2;5:46. doi: 10.1186/1471-2180-5-46. BMC Microbiol. 2005. PMID: 16076392 Free PMC article.
-
Sterol synthesis is essential for viability in the planctomycete bacterium Gemmata obscuriglobus.FEMS Microbiol Lett. 2019 Feb 1;366(3):fnz019. doi: 10.1093/femsle/fnz019. FEMS Microbiol Lett. 2019. PMID: 30715321
-
Lord of the rings--the mechanism for oxidosqualene:lanosterol cyclase becomes crystal clear.Trends Pharmacol Sci. 2005 Jul;26(7):335-40. doi: 10.1016/j.tips.2005.05.004. Trends Pharmacol Sci. 2005. PMID: 15951028 Review.
-
Design strategies of oxidosqualene cyclase inhibitors: Targeting the sterol biosynthetic pathway.J Steroid Biochem Mol Biol. 2017 Jul;171:305-317. doi: 10.1016/j.jsbmb.2017.05.002. Epub 2017 May 4. J Steroid Biochem Mol Biol. 2017. PMID: 28479228 Review.
Cited by
-
Comparative analysis of sterol acquisition in the oomycetes Saprolegnia parasitica and Phytophthora infestans.PLoS One. 2017 Feb 2;12(2):e0170873. doi: 10.1371/journal.pone.0170873. eCollection 2017. PLoS One. 2017. PMID: 28152045 Free PMC article.
-
Genome-wide identification of oxidosqualene cyclase genes regulating natural rubber in Taraxacum kok-saghyz.Planta. 2024 Sep 10;260(4):88. doi: 10.1007/s00425-024-04522-y. Planta. 2024. PMID: 39251530
-
The Role of Gene Duplication in the Divergence of Enzyme Function: A Comparative Approach.Front Genet. 2021 Jul 14;12:641817. doi: 10.3389/fgene.2021.641817. eCollection 2021. Front Genet. 2021. PMID: 34335678 Free PMC article.
-
Genome-Wide Analysis of Oxidosqualene Cyclase Genes in Artemisia annua: Evolution, Expression, and Potential Roles in Triterpenoid Biosynthesis.Curr Issues Mol Biol. 2025 Jul 14;47(7):545. doi: 10.3390/cimb47070545. Curr Issues Mol Biol. 2025. PMID: 40729014 Free PMC article.
References
-
- Abe I., Rohmer M., Prestwich G. D. (1993). Enzymatic cyclization of squalene and oxidosqualene to sterols and triterpenes. Chem. Rev. 93 2189–2206. 10.1021/cr00022a009 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

