Institutional implementation of clinical tumor profiling on an unselected cancer population
- PMID: 27882345
- PMCID: PMC5111542
- DOI: 10.1172/jci.insight.87062
Institutional implementation of clinical tumor profiling on an unselected cancer population
Abstract
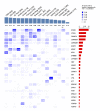
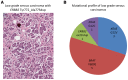
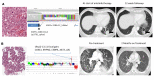
BACKGROUND. Comprehensive genomic profiling of a patient's cancer can be used to diagnose, monitor, and recommend treatment. Clinical implementation of tumor profiling in an enterprise-wide, unselected cancer patient population has yet to be reported. METHODS. We deployed a hybrid-capture and massively parallel sequencing assay (OncoPanel) for all adult and pediatric patients at our combined cancer centers. Results were categorized by pathologists based on actionability. We report the results for the first 3,727 patients tested. RESULTS. Our cohort consists of cancer patients unrestricted by disease site or stage. Across all consented patients, half had sufficient and available (>20% tumor) material for profiling; once specimens were received in the laboratory for pathology review, 73% were scored as adequate for genomic testing. When sufficient DNA was obtained, OncoPanel yielded a result in 96% of cases. 73% of patients harbored an actionable or informative alteration; only 19% of these represented a current standard of care for therapeutic stratification. The findings recapitulate those of previous studies of common cancers but also identify alterations, including in AXL and EGFR, associated with response to targeted therapies. In rare cancers, potentially actionable alterations suggest the utility of a "cancer-agnostic" approach in genomic profiling. Retrospective analyses uncovered contextual genomic features that may inform therapeutic response and examples where diagnoses revised by genomic profiling markedly changed clinical management. CONCLUSIONS. Broad sequencing-based testing deployed across an unselected cancer cohort is feasible. Genomic results may alter management in diverse scenarios; however, additional barriers must be overcome to enable precision cancer medicine on a large scale. FUNDING. This work was supported by DFCI, BWH, and the National Cancer Institute (5R33CA155554 and 5K23CA157631).
Conflict of interest statement
M. Nishino is a consultant for Bristol-Myers Squibb. G.R. Oxnard is a consultant for AstraZeneca, Ariad, and Boehringer-Ingelheim and has honoraria from AstraZeneca and Chugai. L.M. Sholl is a consultant for Genentech. M. Meyerson has research funding from Bayer and a patent on the use of EGFR mutation analysis for lung cancer diagnosis and is a founder of Foundation Medicine. L.A. Garraway is a founder, consultant, and equity holder in Foundation Medicine.
Figures



References
-
- Cobleigh MA, et al. Multinational study of the efficacy and safety of humanized anti-HER2 monoclonal antibody in women who have HER2-overexpressing metastatic breast cancer that has progressed after chemotherapy for metastatic disease. J Clin Oncol. 1999;17(9):2639–2648. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

