Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency
- PMID: 27883040
- PMCID: PMC5121592
- DOI: 10.1038/srep37674
Effects of MeJA on Arabidopsis metabolome under endogenous JA deficiency
Abstract
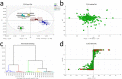
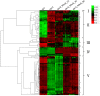
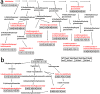
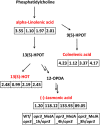
Jasmonates (JAs) play important roles in plant growth, development and defense. Comprehensive metabolomics profiling of plants under JA treatment provides insights into the interaction and regulation network of plant hormones. Here we applied high resolution mass spectrometry based metabolomics approach on Arabidopsis wild type and JA synthesis deficiency mutant opr3. The effects of exogenous MeJA treatment on the metabolites of opr3 were investigated. More than 10000 ion signals were detected and more than 2000 signals showed significant variation in different genotypes and treatment groups. Multivariate statistic analyses (PCA and PLS-DA) were performed and a differential compound library containing 174 metabolites with high resolution precursor ion-product ions pairs was obtained. Classification and pathway analysis of 109 identified compounds in this library showed that glucosinolates and tryptophan metabolism, amino acids and small peptides metabolism, lipid metabolism, especially fatty acyls metabolism, were impacted by endogenous JA deficiency and exogenous MeJA treatment. These results were further verified by quantitative reverse transcription PCR (RT-qPCR) analysis of 21 related genes involved in the metabolism of glucosinolates, tryptophan and α-linolenic acid pathways. The results would greatly enhance our understanding of the biological functions of JA.
Figures

Similar articles
-
Molecular reprogramming of Arabidopsis in response to perturbation of jasmonate signaling.J Proteome Res. 2014 Dec 5;13(12):5751-66. doi: 10.1021/pr500739v. Epub 2014 Oct 29. J Proteome Res. 2014. PMID: 25311705
-
iTRAQ-Based Quantitative Proteomic Analysis of the Arabidopsis Mutant opr3-1 in Response to Exogenous MeJA.Int J Mol Sci. 2020 Jan 16;21(2):571. doi: 10.3390/ijms21020571. Int J Mol Sci. 2020. PMID: 31963133 Free PMC article.
-
Proteomic identification of differentially expressed proteins in Arabidopsis in response to methyl jasmonate.J Plant Physiol. 2011 Jul 1;168(10):995-1008. doi: 10.1016/j.jplph.2011.01.018. Epub 2011 Mar 5. J Plant Physiol. 2011. PMID: 21377756
-
Fatty acid-derived signals in plants.Trends Plant Sci. 2002 May;7(5):217-24. doi: 10.1016/s1360-1385(02)02250-1. Trends Plant Sci. 2002. PMID: 11992827 Review.
-
Transcriptional machineries in jasmonate-elicited plant secondary metabolism.Trends Plant Sci. 2012 Jun;17(6):349-59. doi: 10.1016/j.tplants.2012.03.001. Epub 2012 Mar 27. Trends Plant Sci. 2012. PMID: 22459758 Review.
Cited by
-
Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize.Nat Plants. 2022 Oct;8(10):1176-1190. doi: 10.1038/s41477-022-01254-3. Epub 2022 Oct 14. Nat Plants. 2022. PMID: 36241735
-
Genome-Wide Identification of the Pectate Lyase Gene Family in Potato and Expression Analysis under Salt Stress.Plants (Basel). 2024 May 11;13(10):1322. doi: 10.3390/plants13101322. Plants (Basel). 2024. PMID: 38794393 Free PMC article.
-
Combined Metabolite and Transcriptome Profiling Reveals the Norisoprenoid Responses in Grape Berries to Abscisic Acid and Synthetic Auxin.Int J Mol Sci. 2021 Jan 31;22(3):1420. doi: 10.3390/ijms22031420. Int J Mol Sci. 2021. PMID: 33572582 Free PMC article.
-
Genome-wide identification of the Sec14 gene family and the response to salt and drought stress in soybean (Glycine max).BMC Genomics. 2025 Jan 25;26(1):73. doi: 10.1186/s12864-025-11270-0. BMC Genomics. 2025. PMID: 39863853 Free PMC article.
-
Integrated omics data of two annual ryegrass (Lolium multiflorum L.) genotypes reveals core metabolic processes under drought stress.BMC Plant Biol. 2018 Jan 30;18(1):26. doi: 10.1186/s12870-018-1239-z. BMC Plant Biol. 2018. PMID: 29378511 Free PMC article.
References
-
- Lenz E. M. & Wilson I. D. Analytical strategies in metabonomics. J. Proteome Res. 6, 443–458 (2007). - PubMed
-
- Blasco H. et al.. Metabolomics in cerebrospinal fluid of patients with amyotrophic lateral sclerosis: an untargeted approach via high-resolution mass spectrometry. J. Proteome Res. 12, 3746–3754 (2013). - PubMed
-
- Horn P. J. & Chapman K. D. Lipidomics in situ: insights into plant lipid metabolism from high resolution spatial maps of metabolites. Prog. Lipid Res. 54, 32–52 (2014). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

