Non-RVD mutations that enhance the dynamics of the TAL repeat array along the superhelical axis improve TALEN genome editing efficacy
- PMID: 27883072
- PMCID: PMC5121632
- DOI: 10.1038/srep37887
Non-RVD mutations that enhance the dynamics of the TAL repeat array along the superhelical axis improve TALEN genome editing efficacy
Abstract
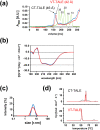
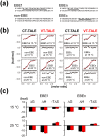
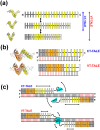
Transcription activator-like effector (TALE) nuclease (TALEN) is widely used as a tool in genome editing. The DNA binding part of TALEN consists of a tandem array of TAL-repeats that form a right-handed superhelix. Each TAL-repeat recognises a specific base by the repeat variable diresidue (RVD) at positions 12 and 13. TALEN comprising the TAL-repeats with periodic mutations to residues at positions 4 and 32 (non-RVD sites) in each repeat (VT-TALE) exhibits increased efficacy in genome editing compared with a counterpart without the mutations (CT-TALE). The molecular basis for the elevated efficacy is unknown. In this report, comparison of the physicochemical properties between CT- and VT-TALEs revealed that VT-TALE has a larger amplitude motion along the superhelical axis (superhelical motion) compared with CT-TALE. The greater superhelical motion in VT-TALE enabled more TAL-repeats to engage in the target sequence recognition compared with CT-TALE. The extended sequence recognition by the TAL-repeats improves site specificity with limiting the spatial distribution of FokI domains to facilitate their dimerization at the desired site. Molecular dynamics simulations revealed that the non-RVD mutations alter inter-repeat hydrogen bonding to amplify the superhelical motion of VT-TALE. The TALEN activity is associated with the inter-repeat hydrogen bonding among the TAL repeats.
Figures


References
-
- Gu K. et al.. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125 (2005). - PubMed
-
- Kay S., Hahn S., Marois E., Hause G. & Bonas U. A Bacterial Effector Acts as a Plant Transcription Factor and Induces a Cell Size Regulator. Science 318, 648–651 (2007). - PubMed
-
- Romer P. et al.. Plant Pathogen Recognition Mediated by Promoter Activation of the Pepper Bs3 Resistance Gene. Science 318, 645–648 (2007). - PubMed
-
- Scholze H. & Boch J. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 14, 47–53 (2011). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

