A CRISPR-Cas9 Assisted Non-Homologous End-Joining Strategy for One-step Engineering of Bacterial Genome
- PMID: 27883076
- PMCID: PMC5121644
- DOI: 10.1038/srep37895
A CRISPR-Cas9 Assisted Non-Homologous End-Joining Strategy for One-step Engineering of Bacterial Genome
Abstract
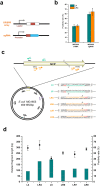
Homologous recombination-mediated genome engineering has been broadly applied in prokaryotes with high efficiency and accuracy. However, this method is limited in realizing larger-scale genome editing with numerous genes or large DNA fragments because of the relatively complicated procedure for DNA editing template construction. Here, we describe a CRISPR-Cas9 assisted non-homologous end-joining (CA-NHEJ) strategy for the rapid and efficient inactivation of bacterial gene (s) in a homologous recombination-independent manner and without the use of selective marker. Our study show that CA-NHEJ can be used to delete large chromosomal DNA fragments in a single step that does not require homologous DNA template. It is thus a novel and powerful tool for bacterial genomes reducing and possesses the potential for accelerating the genome evolution.
Figures


Similar articles
-
An efficient system for deletion of large DNA fragments in Escherichia coli via introduction of both Cas9 and the non-homologous end joining system from Mycobacterium smegmatis.Biochem Biophys Res Commun. 2017 Apr 15;485(4):768-774. doi: 10.1016/j.bbrc.2017.02.129. Epub 2017 Feb 28. Biochem Biophys Res Commun. 2017. PMID: 28257845
-
CRISPR-Cas9-assisted native end-joining editing offers a simple strategy for efficient genetic engineering in Escherichia coli.Appl Microbiol Biotechnol. 2019 Oct;103(20):8497-8509. doi: 10.1007/s00253-019-10104-w. Epub 2019 Sep 9. Appl Microbiol Biotechnol. 2019. PMID: 31501938
-
Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining.Nat Biotechnol. 2015 May;33(5):538-42. doi: 10.1038/nbt.3190. Epub 2015 Mar 23. Nat Biotechnol. 2015. PMID: 25798939 Free PMC article.
-
CRISPR with a Happy Ending: Non-Templated DNA Repair for Prokaryotic Genome Engineering.Biotechnol J. 2020 Jul;15(7):e1900404. doi: 10.1002/biot.201900404. Epub 2020 Jun 29. Biotechnol J. 2020. PMID: 32558098 Review.
-
Strategies for Applying Nonhomologous End Joining-Mediated Genome Editing in Prokaryotes.ACS Synth Biol. 2019 Oct 18;8(10):2194-2202. doi: 10.1021/acssynbio.9b00179. Epub 2019 Sep 26. ACS Synth Biol. 2019. PMID: 31525995 Review.
Cited by
-
Development of an Efficient Genome Editing Tool in Bacillus licheniformis Using CRISPR-Cas9 Nickase.Appl Environ Microbiol. 2018 Mar 1;84(6):e02608-17. doi: 10.1128/AEM.02608-17. Print 2018 Mar 15. Appl Environ Microbiol. 2018. PMID: 29330178 Free PMC article.
-
CRISPR/Cas9-Mediated Customizing Strategies for Adoptive T-Cell Therapy.Pharmaceutics. 2024 Mar 1;16(3):346. doi: 10.3390/pharmaceutics16030346. Pharmaceutics. 2024. PMID: 38543240 Free PMC article. Review.
-
Breeding for water-use efficiency in wheat: progress, challenges and prospects.Mol Biol Rep. 2024 Mar 22;51(1):429. doi: 10.1007/s11033-024-09345-4. Mol Biol Rep. 2024. PMID: 38517566 Review.
-
Nano Approaches to Nucleic Acid Delivery: Barriers, Solutions, and Current Landscape.Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2025 Mar-Apr;17(2):e70010. doi: 10.1002/wnan.70010. Wiley Interdiscip Rev Nanomed Nanobiotechnol. 2025. PMID: 40223402 Free PMC article. Review.
-
Endogenous CRISPR-assisted microhomology-mediated end joining enables rapid genome editing in Zymomonas mobilis.Biotechnol Biofuels. 2021 Oct 24;14(1):208. doi: 10.1186/s13068-021-02056-z. Biotechnol Biofuels. 2021. PMID: 34689795 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

