Structural features of DNA that determine RNA polymerase II core promoter
- PMID: 27884105
- PMCID: PMC5123417
- DOI: 10.1186/s12864-016-3292-z
Structural features of DNA that determine RNA polymerase II core promoter
Abstract
Background: The general structure and action of all eukaryotic and archaeal RNA polymerases machinery have an astonishing similarity despite the diversity of core promoter sequences in different species. The goal of our work is to find common characteristics of DNA region that define it as a promoter for the RNA polymerase II (Pol II).
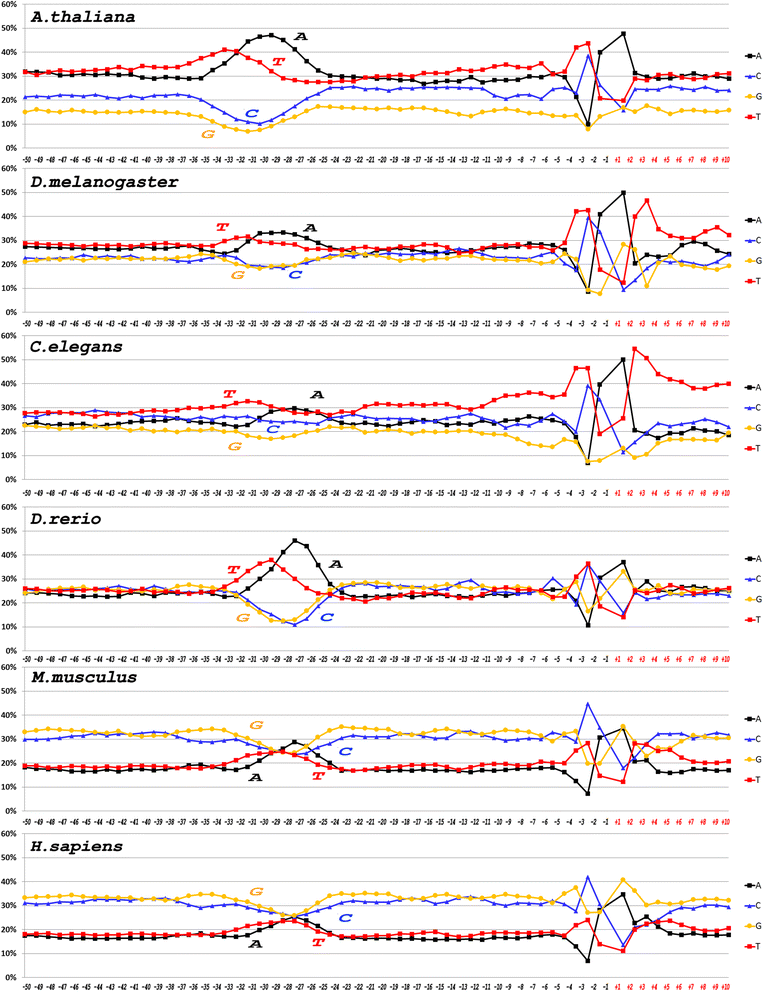
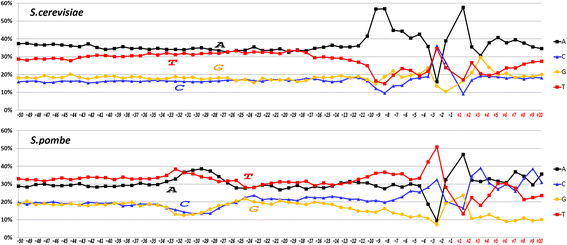
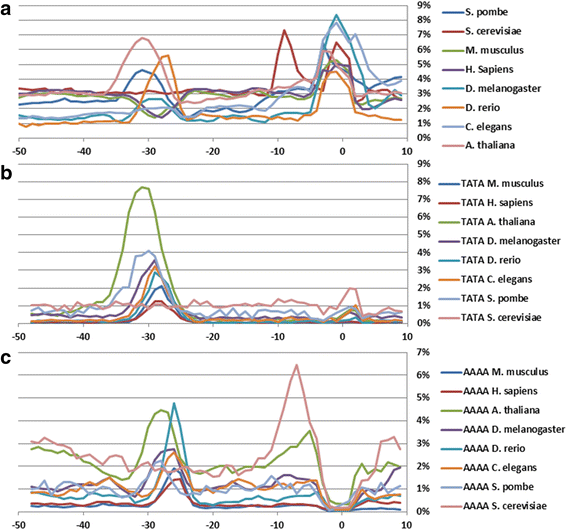
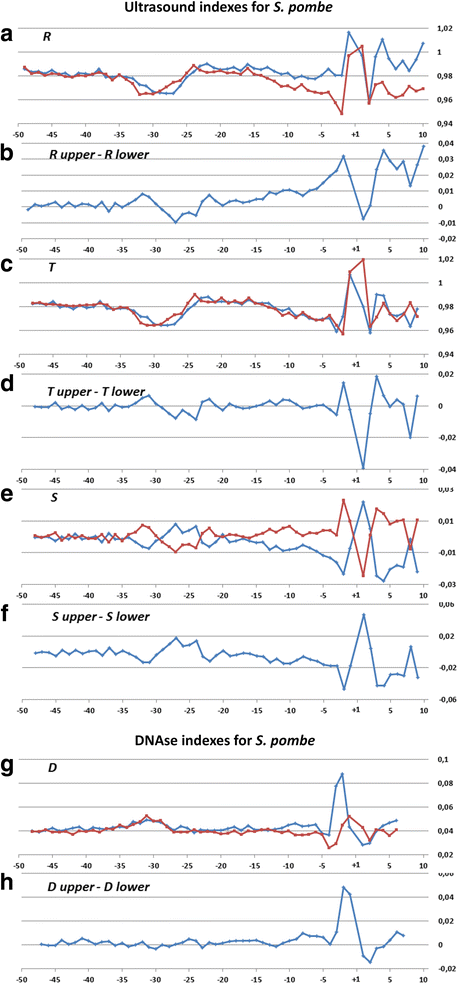
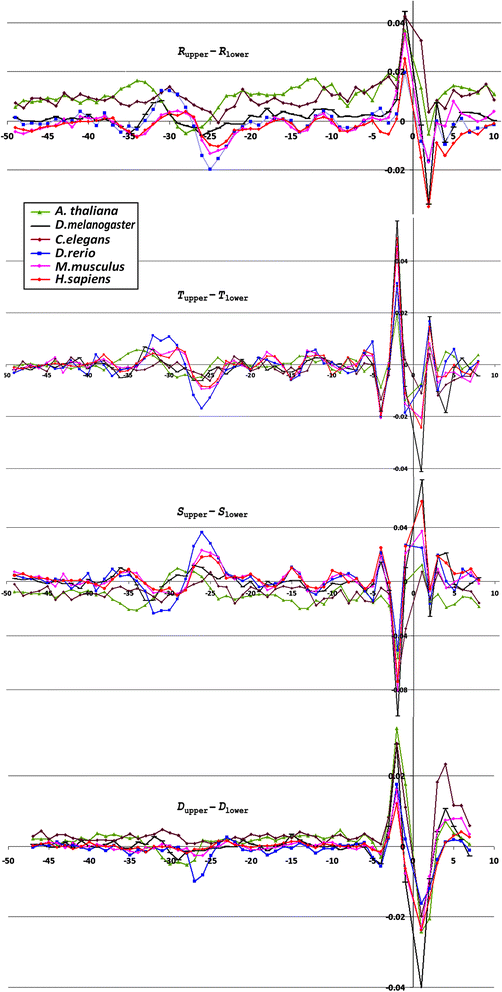
Results: The profiles of a large number of physical and structural characteristics, averaged over representative sets of the Pol II minimal core promoters of the evolutionary divergent species from animals, plants and unicellular fungi were analysed. In addition to the characteristics defined at the base-pair steps, we, for the first time, use profiles of the ultrasonic cleavage and DNase I cleavage indexes, informative for internal properties of each complementary strand.
Conclusions: DNA of the core promoters of metazoans and Schizosaccharomyces pombe has similar structural organization. Its mechanical and 3D structural characteristics have singular properties at the positions of TATA-box. The minor groove is broadened and conformational motion is decreased in that region. Special characteristics of conformational behavior are revealed in metazoans at the region, which connects the end of TATA-box and the transcription start site (TSS). The intensities of conformational motions in the complementary strands are periodically changed in opposite phases. They are noticeable, best of all, in mammals. Such conformational features are lacking in the core promoters of S. pombe. The profiles of Saccharomyces cerevisiae core promoters significantly differ: their singular region is shifted down thus pointing to the uniqueness of their structural organization. Obtained results may be useful in genetic engineering for artificial modulation of the promoter strength.
Keywords: Local DNA structure; RNA polymerase II promoter sequences; Sequence-specific ultrasonic cleavage.
Figures







Similar articles
-
Evolutionary Invariant of the Structure of DNA Double Helix in RNAP II Core Promoters.Int J Mol Sci. 2022 Sep 17;23(18):10873. doi: 10.3390/ijms231810873. Int J Mol Sci. 2022. PMID: 36142782 Free PMC article.
-
Genome-wide analysis of core promoter structures in Schizosaccharomyces pombe with DeepCAGE.RNA Biol. 2015;12(5):525-37. doi: 10.1080/15476286.2015.1022704. RNA Biol. 2015. PMID: 25747261 Free PMC article.
-
Widespread use of TATA elements in the core promoters for RNA polymerases III, II, and I in fission yeast.Mol Cell Biol. 2001 Oct;21(20):6870-81. doi: 10.1128/MCB.21.20.6870-6881.2001. Mol Cell Biol. 2001. PMID: 11564871 Free PMC article.
-
RNA polymerase II transcription apparatus in Schizosaccharomyces pombe.Curr Genet. 2004 Jan;44(6):287-94. doi: 10.1007/s00294-003-0446-8. Epub 2003 Oct 22. Curr Genet. 2004. PMID: 14574615 Review.
-
Perspectives on the RNA polymerase II core promoter.Wiley Interdiscip Rev Dev Biol. 2012 Jan-Feb;1(1):40-51. doi: 10.1002/wdev.21. Epub 2011 Dec 6. Wiley Interdiscip Rev Dev Biol. 2012. PMID: 23801666 Free PMC article. Review.
Cited by
-
A Method for Identification of the Methylation Level of CpG Islands From NGS Data.Sci Rep. 2020 May 25;10(1):8635. doi: 10.1038/s41598-020-65406-1. Sci Rep. 2020. PMID: 32451390 Free PMC article.
-
Structural Features of DNA in tRNA Genes and Their Upstream Sequences.Int J Mol Sci. 2024 Nov 1;25(21):11758. doi: 10.3390/ijms252111758. Int J Mol Sci. 2024. PMID: 39519309 Free PMC article.
-
TSSFinder-fast and accurate ab initio prediction of the core promoter in eukaryotic genomes.Brief Bioinform. 2021 Nov 5;22(6):bbab198. doi: 10.1093/bib/bbab198. Brief Bioinform. 2021. PMID: 34050351 Free PMC article.
-
Evolutionary Invariant of the Structure of DNA Double Helix in RNAP II Core Promoters.Int J Mol Sci. 2022 Sep 17;23(18):10873. doi: 10.3390/ijms231810873. Int J Mol Sci. 2022. PMID: 36142782 Free PMC article.
-
Initiator-Directed Transcription: Fission Yeast Nmtl Initiator Directs Preinitiation Complex Formation and Transcriptional Initiation.Genes (Basel). 2022 Jan 28;13(2):256. doi: 10.3390/genes13020256. Genes (Basel). 2022. PMID: 35205301 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

