Patterns of ribosomal protein expression specify normal and malignant human cells
- PMID: 27884178
- PMCID: PMC5123215
- DOI: 10.1186/s13059-016-1104-z
Patterns of ribosomal protein expression specify normal and malignant human cells
Abstract
Background: Ribosomes are highly conserved molecular machines whose core composition has traditionally been regarded as invariant. However, recent studies have reported intriguing differences in the expression of some ribosomal proteins (RPs) across tissues and highly specific effects on the translation of individual mRNAs.
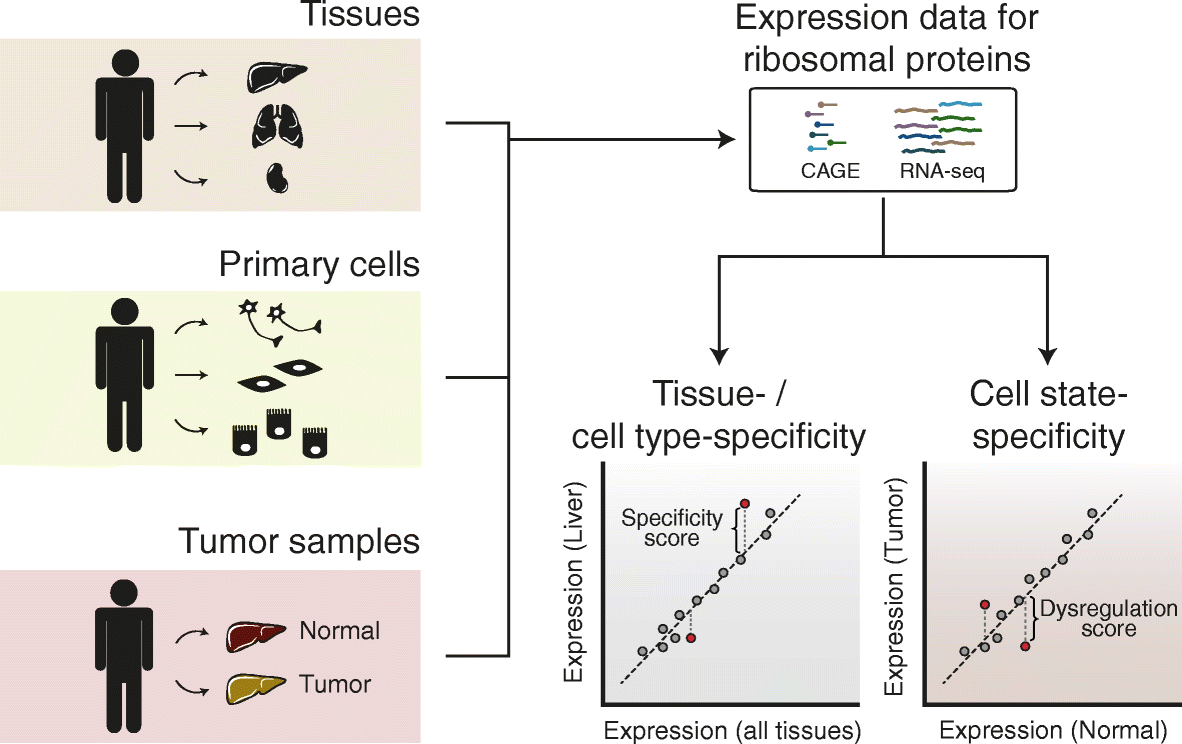
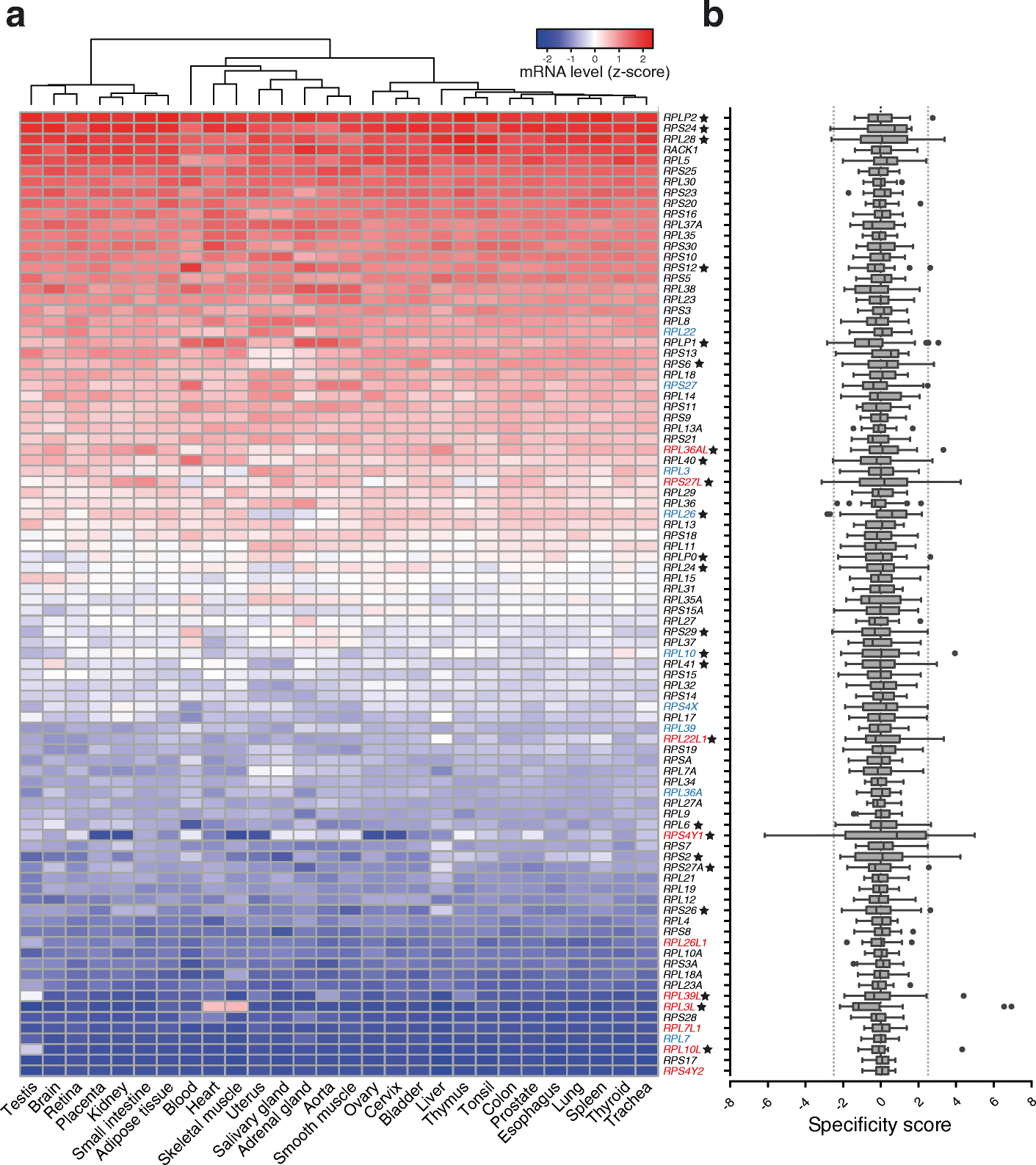
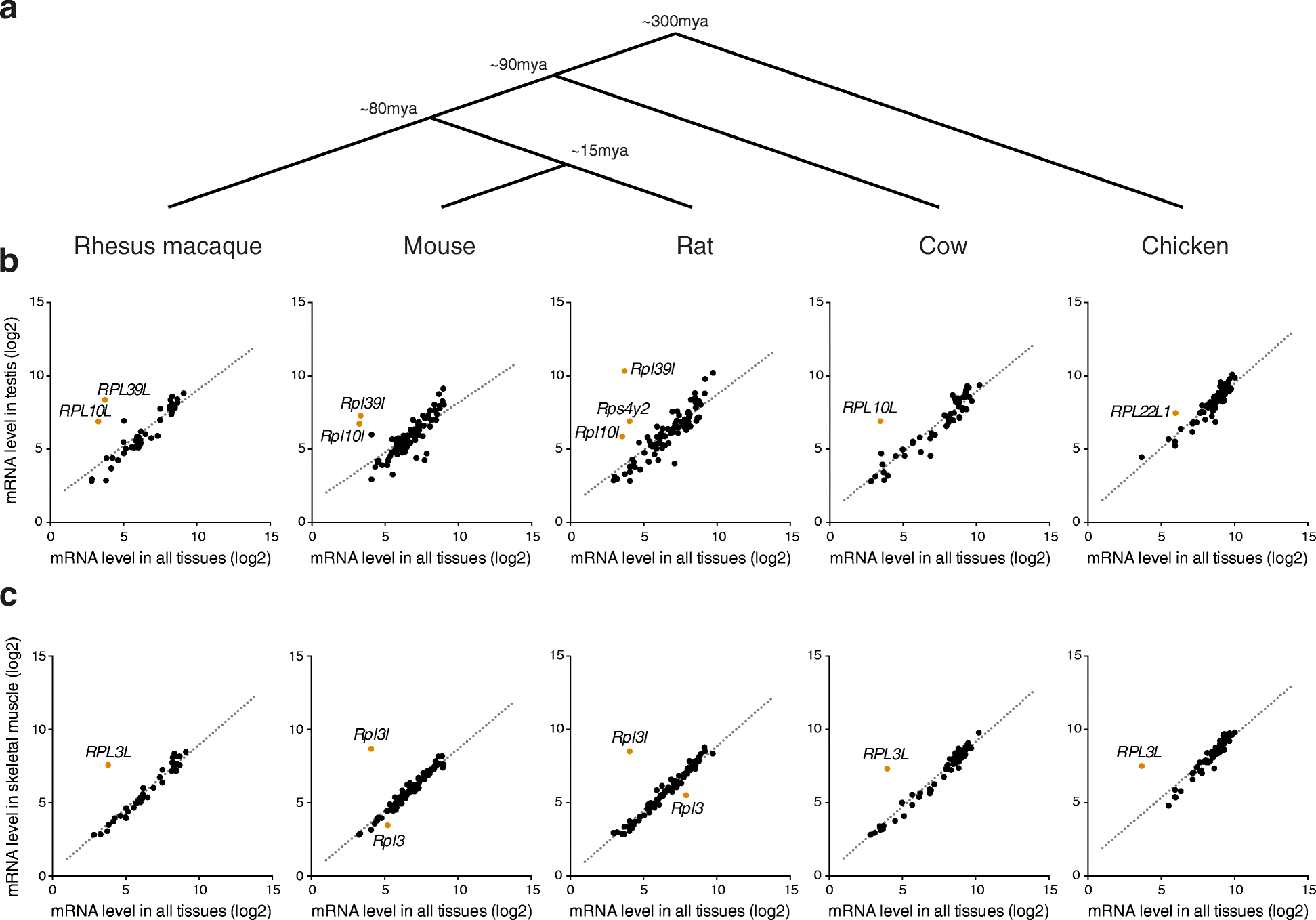
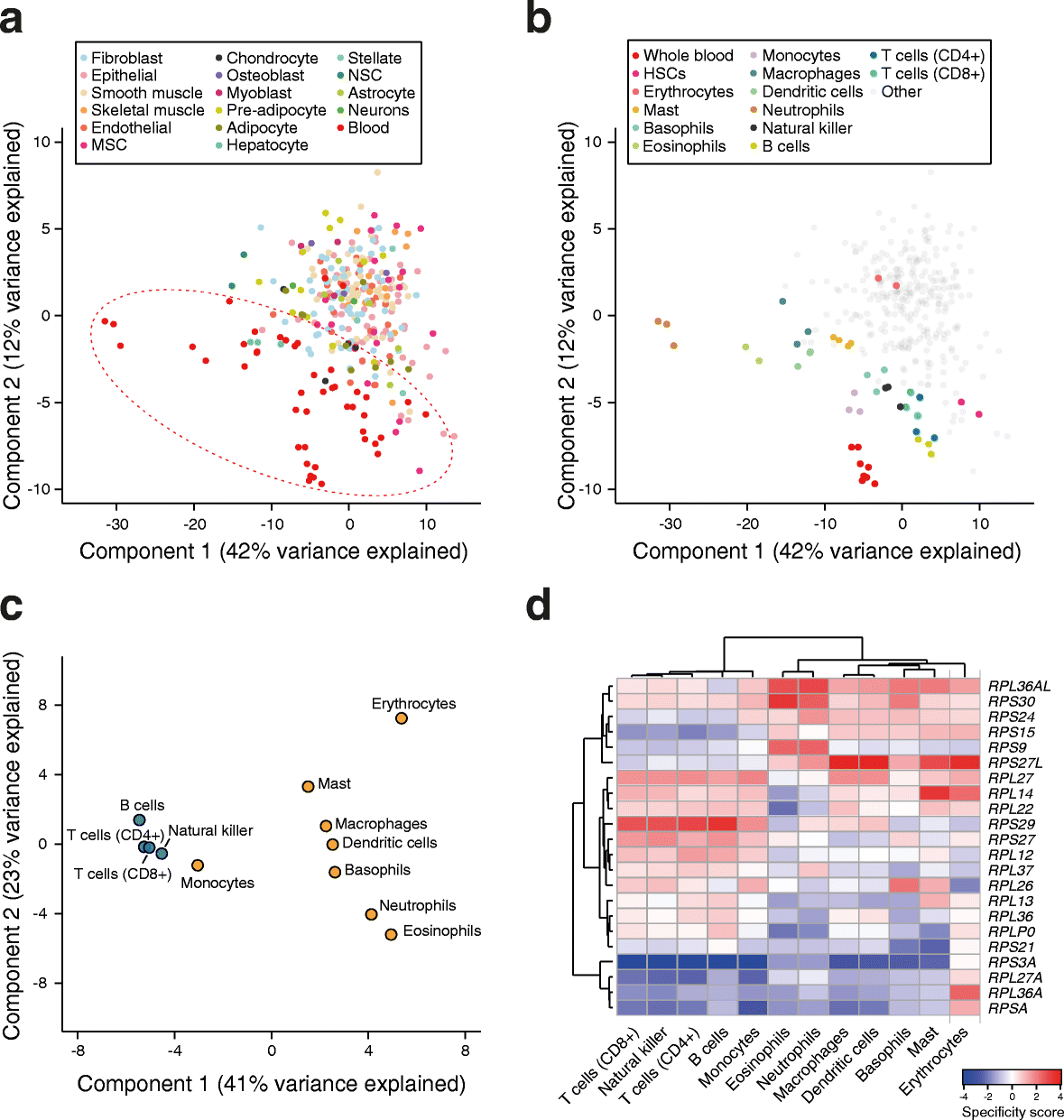
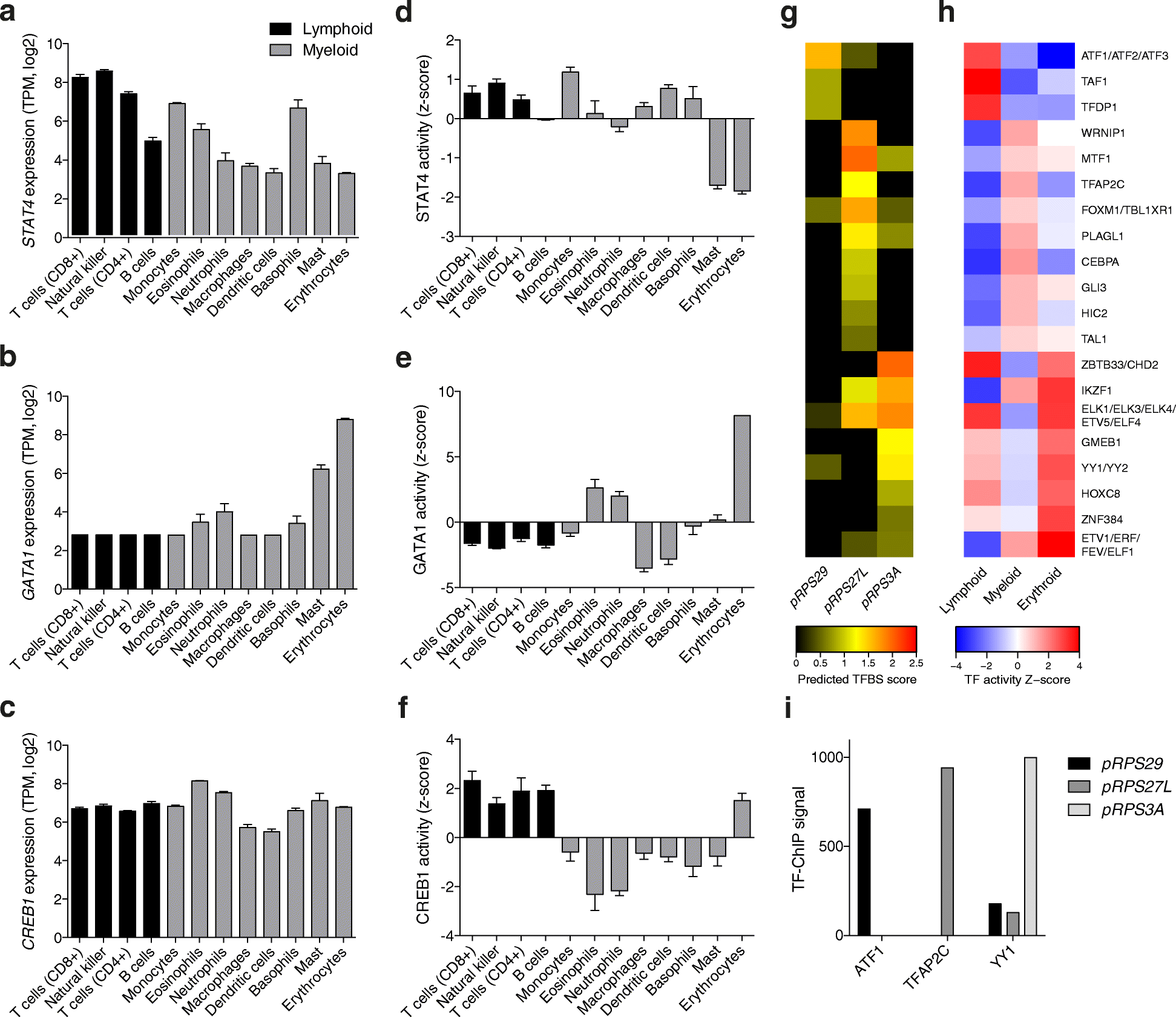
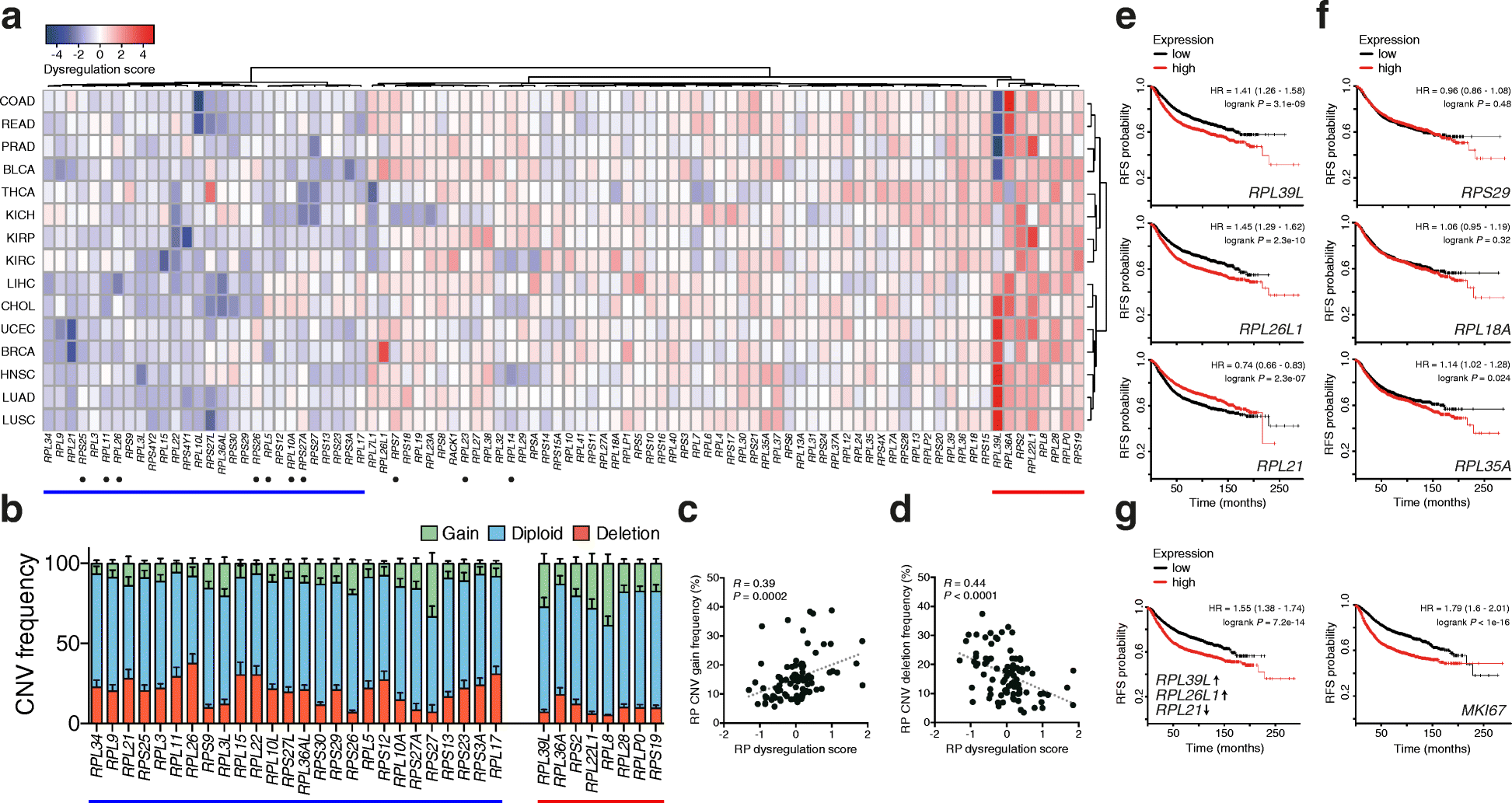
Results: To determine whether RPs are more generally linked to cell identity, we analyze the heterogeneity of RP expression in a large set of human tissues, primary cells, and tumors. We find that about a quarter of human RPs exhibit tissue-specific expression and that primary hematopoietic cells display the most complex patterns of RP expression, likely shaped by context-restricted transcriptional regulators. Strikingly, we uncover patterns of dysregulated expression of individual RPs across cancer types that arise through copy number variations and are predictive for disease progression.
Conclusions: Our study reveals an unanticipated plasticity of RP expression across normal and malignant human cell types and provides a foundation for future characterization of cellular behaviors that are orchestrated by specific RPs.
Keywords: Cancer; Hematopoiesis; Ribosomal proteins; Ribosome heterogeneity; Translation.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

