Aldehyde dehydrogenase is used by cancer cells for energy metabolism
- PMID: 27885254
- PMCID: PMC5133370
- DOI: 10.1038/emm.2016.103
Aldehyde dehydrogenase is used by cancer cells for energy metabolism
Abstract
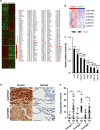
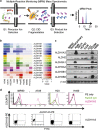
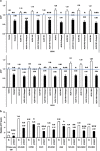
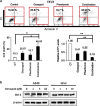
We found that non-small-cell lung cancer (NSCLC) cells express high levels of multiple aldehyde dehydrogenase (ALDH) isoforms via an informatics analysis of metabolic enzymes in NSCLC and immunohistochemical staining of NSCLC clinical tumor samples. Using a multiple reaction-monitoring mass spectrometry analysis, we found that multiple ALDH isozymes were generally abundant in NSCLC cells compared with their levels in normal IMR-90 human lung cells. As a result of the catalytic reaction mediated by ALDH, NADH is produced as a by-product from the conversion of aldehyde to carboxylic acid. We hypothesized that the NADH produced by ALDH may be a reliable energy source for ATP production in NSCLC. This study revealed that NADH production by ALDH contributes significantly to ATP production in NSCLC. Furthermore, gossypol, a pan-ALDH inhibitor, markedly reduced the level of ATP. Gossypol combined with phenformin synergistically reduced the ATP levels, which efficiently induced cell death following cell cycle arrest.
Figures

References
-
- Wong DW, Leung EL, So KK, Tam IY, Sihoe AD, Cheng LC et al. The EML4-ALK fusion gene is involved in various histologic types of lung cancers from nonsmokers with wild-type EGFR and KRAS. Cancer 2009; 115: 1723–1733. - PubMed
-
- Li X, Wan L, Geng J, Wu CL, Bai X. Aldehyde dehydrogenase 1A1 possesses stem-like properties and predicts lung cancer patient outcome. J Thorac Oncol 2012; 7: 1235–1245. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

