Mapping out Min protein patterns in fully confined fluidic chambers
- PMID: 27885986
- PMCID: PMC5217063
- DOI: 10.7554/eLife.19271
Mapping out Min protein patterns in fully confined fluidic chambers
Abstract
The bacterial Min protein system provides a major model system for studying reaction-diffusion processes in biology. Here we present the first in vitro study of the Min system in fully confined three-dimensional chambers that are lithography-defined, lipid-bilayer coated and isolated through pressure valves. We identify three typical dynamical behaviors that occur dependent on the geometrical chamber parameters: pole-to-pole oscillations, spiral rotations, and traveling waves. We establish the geometrical selection rules and show that, surprisingly, Min-protein spiral rotations govern the larger part of the geometrical phase diagram. Confinement as well as an elevated temperature reduce the characteristic wavelength of the Min patterns, although even for confined chambers with a bacterial-level viscosity, the patterns retain a ~5 times larger wavelength than in vivo. Our results provide an essential experimental base for modeling of intracellular Min gradients in bacterial cell division as well as, more generally, for understanding pattern formation in reaction-diffusion systems.
Keywords: E. coli; Microfluidics; Min system; biophysics; in vitro reconstitution; pattern formation; reaction-diffusion; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

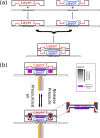




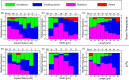


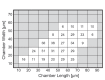




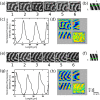
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

