Accelerated cell divisions drive the outgrowth of the regenerating spinal cord in axolotls
- PMID: 27885987
- PMCID: PMC5182066
- DOI: 10.7554/eLife.20357
Accelerated cell divisions drive the outgrowth of the regenerating spinal cord in axolotls
Abstract
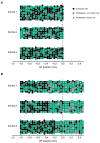
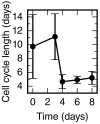
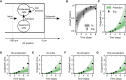
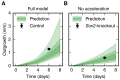
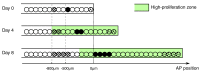
Axolotls are unique in their ability to regenerate the spinal cord. However, the mechanisms that underlie this phenomenon remain poorly understood. Previously, we showed that regenerating stem cells in the axolotl spinal cord revert to a molecular state resembling embryonic neuroepithelial cells and functionally acquire rapid proliferative divisions (Rodrigo Albors et al., 2015). Here, we refine the analysis of cell proliferation in space and time and identify a high-proliferation zone in the regenerating spinal cord that shifts posteriorly over time. By tracking sparsely-labeled cells, we also quantify cell influx into the regenerate. Taking a mathematical modeling approach, we integrate these quantitative datasets of cell proliferation, neural stem cell activation and cell influx, to predict regenerative tissue outgrowth. Our model shows that while cell influx and neural stem cell activation play a minor role, the acceleration of the cell cycle is the major driver of regenerative spinal cord outgrowth in axolotls.
Keywords: axolotl; cell proliferation; computational biology; developmental biology; modeling; regeneration; stem cells; systems biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures







References
-
- D'Agostino RB. An omnibus test of normality for moderate and large size samples. Biometrika. 1971;58:341–348. doi: 10.1093/biomet/58.2.341. - DOI
-
- D’Agostino R, Pearson ES. Tests for departure from normality. Empirical results for the distributions of b2 and √b1. Biometrika. 1973;60:613–622. doi: 10.1093/biomet/60.3.613. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

