Histone deacetylase 1 plays a predominant pro-oncogenic role in Eμ-myc driven B cell lymphoma
- PMID: 27886239
- PMCID: PMC5122906
- DOI: 10.1038/srep37772
Histone deacetylase 1 plays a predominant pro-oncogenic role in Eμ-myc driven B cell lymphoma
Abstract
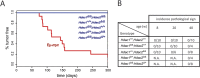
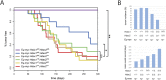
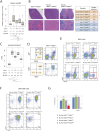
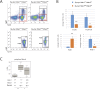
The two histone deacetylases (Hdacs), Hdac1 and Hdac2, are erasers of acetylation marks on histone tails, and are important regulators of gene expression that were shown to play important roles in hematological malignancies. However, several recent studies reported opposing tumor-suppressive or tumor-promoting roles for Hdac1 and Hdac2. Here, we investigated the functional role of Hdac1 and Hdac2 using the Eμ-myc mouse model of B cell lymphoma. We demonstrate that Hdac1 and Hdac2 have a pro-oncogenic role in both Eμ-myc tumorigenesis and tumor maintenance. Hdac1 and Hdac2 promote tumorigenesis in a gene dose-dependent manner, with a predominant function of Hdac1. Our data show that Hdac1 and Hdac2 impact on Eμ-myc B cell proliferation and apoptosis and suggest that a critical level of Hdac activity may be required for Eμ-myc tumorigenesis and proper B cell development. This provides the rationale for utilization of selective Hdac1 and Hdac2 inhibitors in the treatment of hematological malignancies.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

