An evolutionary conserved interaction between the Gcm transcription factor and the SF1 nuclear receptor in the female reproductive system
- PMID: 27886257
- PMCID: PMC5122895
- DOI: 10.1038/srep37792
An evolutionary conserved interaction between the Gcm transcription factor and the SF1 nuclear receptor in the female reproductive system
Abstract
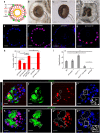
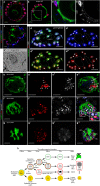
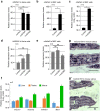
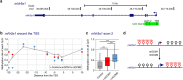
NR5A1 is essential for the development and for the function of steroid producing glands of the reproductive system. Moreover, its misregulation is associated with endometriosis, which is the first cause of infertility in women. Hr39, the Drosophila ortholog of NR5A1, is expressed and required in the secretory cells of the spermatheca, the female exocrine gland that ensures fertility by secreting substances that attract and capacitate the spermatozoids. We here identify a direct regulator of Hr39 in the spermatheca: the Gcm transcription factor. Furthermore, lack of Gcm prevents the production of the secretory cells and leads to female sterility in Drosophila. Hr39 regulation by Gcm seems conserved in mammals and involves the modification of the DNA methylation profile of mNr5a1. This study identifies a new molecular pathway in female reproductive system development and suggests a role for hGCM in the progression of reproductive tract diseases in humans.
Figures


Similar articles
-
The Sf1-related nuclear hormone receptor Hr39 regulates Drosophila female reproductive tract development and function.Development. 2008 Jan;135(2):311-21. doi: 10.1242/dev.015156. Epub 2007 Dec 12. Development. 2008. PMID: 18077584
-
NR5A nuclear receptor Hr39 controls three-cell secretory unit formation in Drosophila female reproductive glands.Curr Biol. 2012 May 22;22(10):862-71. doi: 10.1016/j.cub.2012.03.059. Epub 2012 May 3. Curr Biol. 2012. PMID: 22560612 Free PMC article.
-
Regulation of male fertility and accessory gland gene expression by the Drosophila HR39 nuclear receptor.Dev Biol. 2021 Nov;479:51-60. doi: 10.1016/j.ydbio.2021.07.011. Epub 2021 Jul 29. Dev Biol. 2021. PMID: 34331899 Free PMC article.
-
Evolutionary Conservation of the Gcm/Glide Cascade: Of Glia and Beyond.Brain Behav Evol. 2025;100(1):58-66. doi: 10.1159/000542753. Epub 2024 Nov 25. Brain Behav Evol. 2025. PMID: 39586239 Review.
-
Gcm proteins function in the developing nervous system.Dev Biol. 2012 Oct 1;370(1):63-70. doi: 10.1016/j.ydbio.2012.07.018. Epub 2012 Jul 24. Dev Biol. 2012. PMID: 22842100 Review.
Cited by
-
Transcriptional profiling and physiological roles of Aedes aegypti spermathecal-related genes.BMC Genomics. 2020 Feb 10;21(1):143. doi: 10.1186/s12864-020-6543-y. BMC Genomics. 2020. PMID: 32041546 Free PMC article.
-
Finishing the egg.Genetics. 2024 Jan 3;226(1):iyad183. doi: 10.1093/genetics/iyad183. Genetics. 2024. PMID: 38000906 Free PMC article.
-
Dynamic Notch Signaling Specifies Each Cell Fate in Drosophila Spermathecal Lineage.G3 (Bethesda). 2017 May 5;7(5):1417-1427. doi: 10.1534/g3.117.040212. G3 (Bethesda). 2017. PMID: 28258114 Free PMC article.
-
Different modes of Notch activation and strength regulation in the spermathecal secretory lineage.Development. 2020 Feb 7;147(3):dev184390. doi: 10.1242/dev.184390. Development. 2020. PMID: 31988187 Free PMC article.
-
Butterfly eyespots exhibit unique patterns of open chromatin.F1000Res. 2023 Oct 31;12:1428. doi: 10.12688/f1000research.133789.1. eCollection 2023. F1000Res. 2023. PMID: 38778811 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

