Cancer somatic mutations cluster in a subset of regulatory sites predicted from the ENCODE data
- PMID: 27887606
- PMCID: PMC5123355
- DOI: 10.1186/s12943-016-0560-0
Cancer somatic mutations cluster in a subset of regulatory sites predicted from the ENCODE data
Abstract
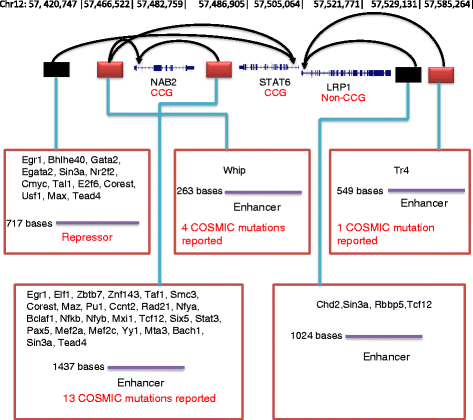
Background: Transcriptional regulation of gene expression is essential for cellular differentiation and function, and defects in the process are associated with cancer. The ENCODE project has mapped potential regulatory sites across the complete genome in many cell types, and these regions have been shown to harbour many of the somatic mutations that occur in cancer cells, suggesting that their effects may drive cancer initiation and development. The ENCODE data suggests a very large number of regulatory sites, and methods are needed to identify those that are most relevant and to connect them to the genes that they control.
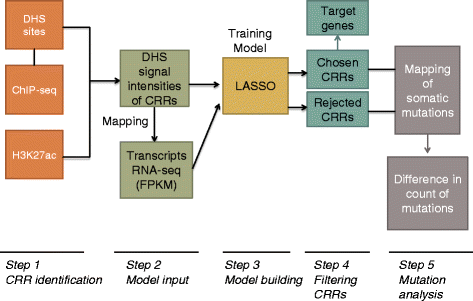
Methods: Predictive models of gene expression were developed by integrating the ENCODE data for regulation, including transcription factor binding and DNase1 hypersensitivity, with RNA-seq data for gene expression. A penalized regression method was used to identify the most predictive potential regulatory sites for each transcript. Known cancer somatic mutations from the COSMIC database were mapped to potential regulatory sites, and we examined differences in the mapping frequencies associated with sites chosen in regulatory models and other (rejected) sites. The effects of potential confounders, for example replication timing, were considered.
Results: Cancer somatic mutations preferentially occupy those regulatory regions chosen in our models as most predictive of gene expression.
Conclusion: Our methods have identified a significantly reduced set of regulatory sites that are enriched in cancer somatic mutations and are more predictive of gene expression. This has significance for the mechanistic interpretation of cancer mutations, and the understanding of genetic regulation.
Keywords: Cancer mutations; Cis regulation; Gene regulation; Modelling; Regulatory regions.
Figures

References
-
- Vinagre J, Almeida A, Populo H, Batista R, Lyra J, Pinto V, Coelho R, Celestino R, Prazeres H, Lima L, et al. Frequency of TERT promoter mutations in human cancers. Nat Commun. 2013;4. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

