Developing a set of strong intronic promoters for robust metabolic engineering in oleaginous Rhodotorula (Rhodosporidium) yeast species
- PMID: 27887615
- PMCID: PMC5124236
- DOI: 10.1186/s12934-016-0600-x
Developing a set of strong intronic promoters for robust metabolic engineering in oleaginous Rhodotorula (Rhodosporidium) yeast species
Abstract
Background: Red yeast species in the Rhodotorula/Rhodosporidium genus are outstanding producers of triacylglyceride and cell biomass. Metabolic engineering is expected to further enhance the productivity and versatility of these hosts for the production of biobased chemicals and fuels. Promoters with strong activity during oil-accumulation stage are critical tools for metabolic engineering of these oleaginous yeasts.
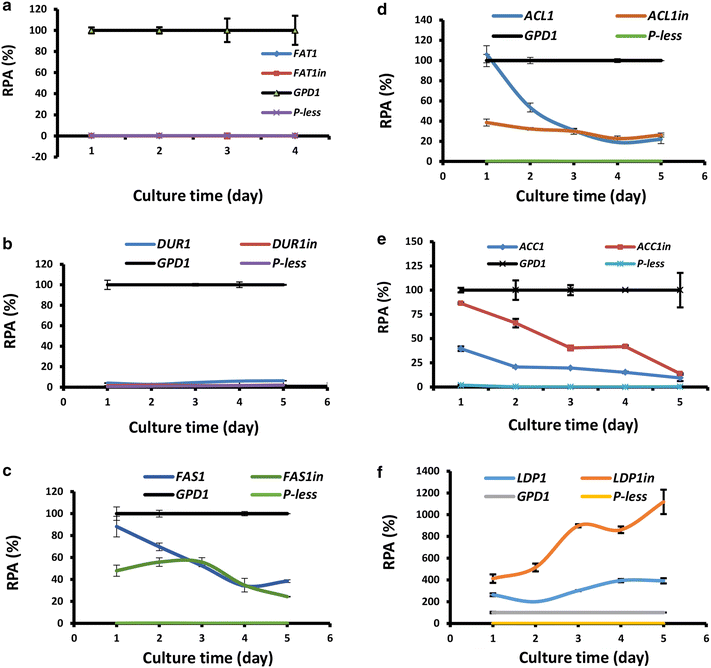
Results: The upstream DNA sequences of 6 genes involved in lipid biosynthesis or accumulation in Rhodotorula toruloides were studied by luciferase reporter assay. The promoter of perilipin/lipid droplet protein 1 gene (LDP1) displayed much stronger activity (4-11 folds) than that of glyceraldehyde-3-phosphate dehydrogenase gene (GPD1), one of the strongest promoters known in yeasts. Depending on the stage of cultivation, promoter of acetyl-CoA carboxylase gene (ACC1) and fatty acid synthase β subunit gene (FAS1) exhibited intermediate strength, displaying 50-160 and 20-90% levels of GPD1 promoter, respectively. Interestingly, introns significantly modulated promoter strength at high frequency. The incorporation of intron 1 and 2 of LDP1 (LDP1in promoter) enhanced its promoter activity by 1.6-3.0 folds. Similarly, the strength of ACC1 promoter was enhanced by 1.5-3.2 folds if containing intron 1. The intron 1 sequences of ACL1 and FAS1 also played significant regulatory roles. When driven by the intronic promoters of ACC1 and LDP1 (ACC1in and LDP1in promoter, respectively), the reporter gene expression were up-regulated by nitrogen starvation, independent of de novo oil biosynthesis and accumulation. As a proof of principle, overexpression of the endogenous acyl-CoA-dependent diacylglycerol acyltransferase 1 gene (DGA1) by LDP1in promoter was significantly more efficient than GPD1 promoter in enhancing lipid accumulation.
Conclusion: Intronic sequences play an important role in regulating gene expression in R. toruloides. Three intronic promoters, LDP1in, ACC1in and FAS1in, are excellent promoters for metabolic engineering in the oleaginous and carotenogenic yeast, R. toruloides.
Keywords: Lipid; Metabolic engineering; Oleaginous yeast; Promoter; Rhodosporidium/Rhodotorula.
Figures




Similar articles
-
Engineering an efficient and tight D-amino acid-inducible gene expression system in Rhodosporidium/Rhodotorula species.Microb Cell Fact. 2015 Oct 26;14:170. doi: 10.1186/s12934-015-0357-7. Microb Cell Fact. 2015. PMID: 26502730 Free PMC article.
-
Metabolic engineering of the oleaginous yeast Rhodosporidium toruloides IFO0880 for lipid overproduction during high-density fermentation.Appl Microbiol Biotechnol. 2016 Nov;100(21):9393-9405. doi: 10.1007/s00253-016-7815-y. Epub 2016 Sep 27. Appl Microbiol Biotechnol. 2016. PMID: 27678117
-
Metabolic engineering of Rhodotorula toruloides IFO0880 improves C16 and C18 fatty alcohol production from synthetic media.Microb Cell Fact. 2022 Feb 19;21(1):26. doi: 10.1186/s12934-022-01750-3. Microb Cell Fact. 2022. PMID: 35183175 Free PMC article.
-
Fatty acids from oleaginous yeasts and yeast-like fungi and their potential applications.Crit Rev Biotechnol. 2018 Nov;38(7):1049-1060. doi: 10.1080/07388551.2018.1428167. Epub 2018 Feb 1. Crit Rev Biotechnol. 2018. PMID: 29385857 Review.
-
Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology.Prog Lipid Res. 2013 Oct;52(4):395-408. doi: 10.1016/j.plipres.2013.05.002. Epub 2013 May 16. Prog Lipid Res. 2013. PMID: 23685199 Review.
Cited by
-
Systems and Synthetic Biology Approaches to Engineer Fungi for Fine Chemical Production.Front Bioeng Biotechnol. 2018 Oct 3;6:117. doi: 10.3389/fbioe.2018.00117. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30338257 Free PMC article. Review.
-
Development of a dedicated Golden Gate Assembly Platform (RtGGA) for Rhodotorula toruloides.Metab Eng Commun. 2022 May 23;15:e00200. doi: 10.1016/j.mec.2022.e00200. eCollection 2022 Dec. Metab Eng Commun. 2022. PMID: 35662893 Free PMC article.
-
A toolset of constitutive promoters for metabolic engineering of Rhodosporidium toruloides.Microb Cell Fact. 2019 Jun 29;18(1):117. doi: 10.1186/s12934-019-1167-0. Microb Cell Fact. 2019. PMID: 31255171 Free PMC article.
-
Impairment of carotenoid biosynthesis through CAR1 gene mutation results in CoQ10, sterols, and phytoene accumulation in Rhodotorula mucilaginosa.Appl Microbiol Biotechnol. 2022 Jan;106(1):317-327. doi: 10.1007/s00253-021-11673-5. Epub 2021 Dec 15. Appl Microbiol Biotechnol. 2022. PMID: 34910239
-
Rhodosporidium toruloides - A potential red yeast chassis for lipids and beyond.FEMS Yeast Res. 2020 Aug 1;20(5):foaa038. doi: 10.1093/femsyr/foaa038. FEMS Yeast Res. 2020. PMID: 32614407 Free PMC article. Review.
References
-
- Sampaio JP, Gadanho M, Bauer R, Weiß M. Taxonomic studies in the Microbotryomycetidae: Leucosporidium golubevii sp. nov., Leucosporidiella gen. nov. and the new orders Leucosporidiales and Sporidiobolales. Mycol Prog. 2003;2:53–68. doi: 10.1007/s11557-006-0044-5. - DOI
-
- Yamazaki M, Komagata K. Taxonomic significance of electrophoretic comparison of enzymes in the genera Rhodotorula and Rhodosporidium. Int J Syst Bacteriol. 1981;31:361–381. doi: 10.1099/00207713-31-3-361. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

