Parameter set for computer-assisted texture analysis of fetal brain
- PMID: 27887658
- PMCID: PMC5124296
- DOI: 10.1186/s13104-016-2300-3
Parameter set for computer-assisted texture analysis of fetal brain
Abstract
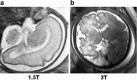
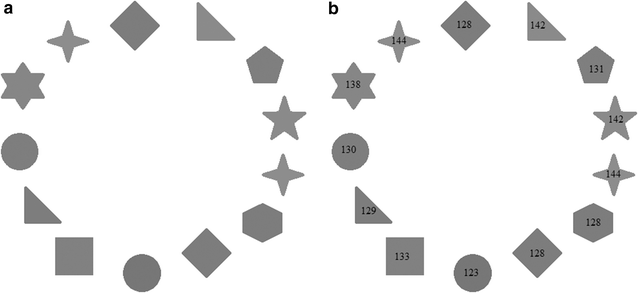
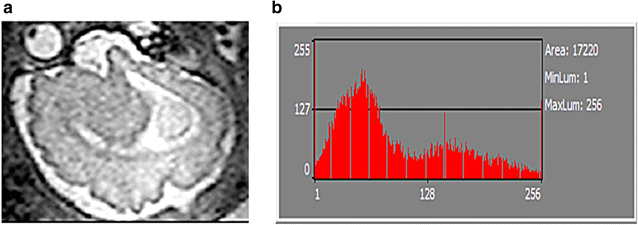
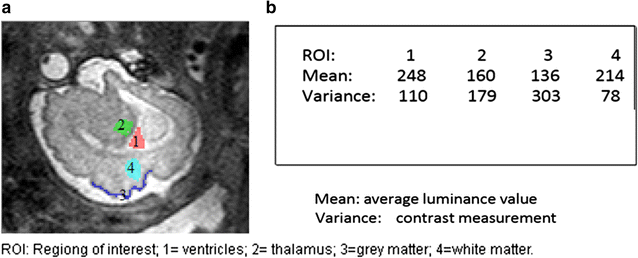
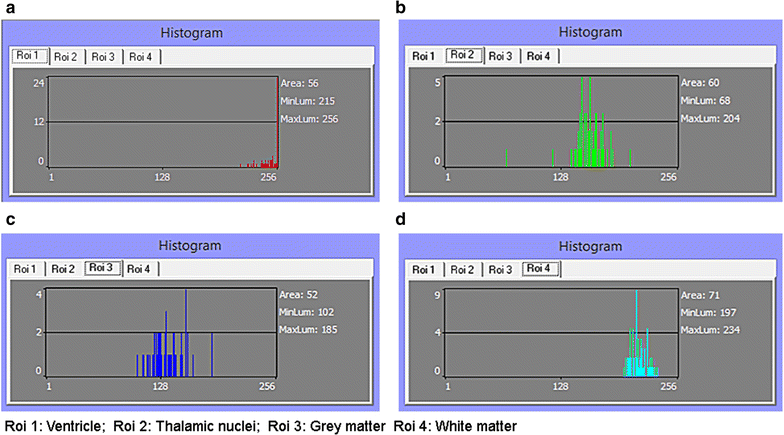
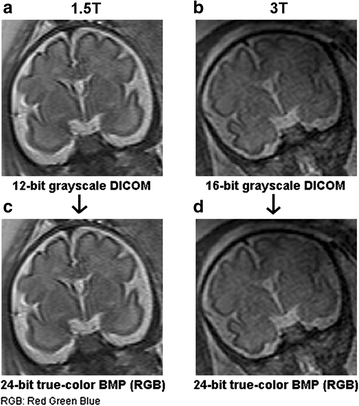
Background: Magnetic resonance data were collected from a diverse population of gravid women to objectively compare the quality of 1.5-tesla (1.5 T) versus 3-T magnetic resonance imaging of the developing human brain. MaZda and B11 computational-visual cognition tools were used to process 2D images. We proposed a wavelet-based parameter and two novel histogram-based parameters for Fisher texture analysis in three-dimensional space.
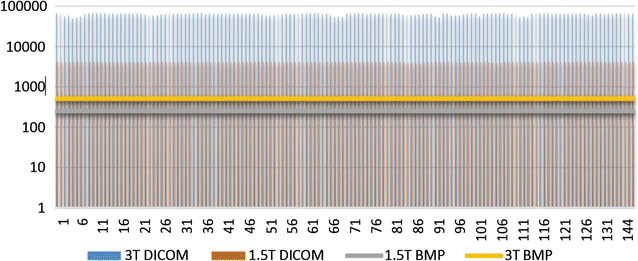
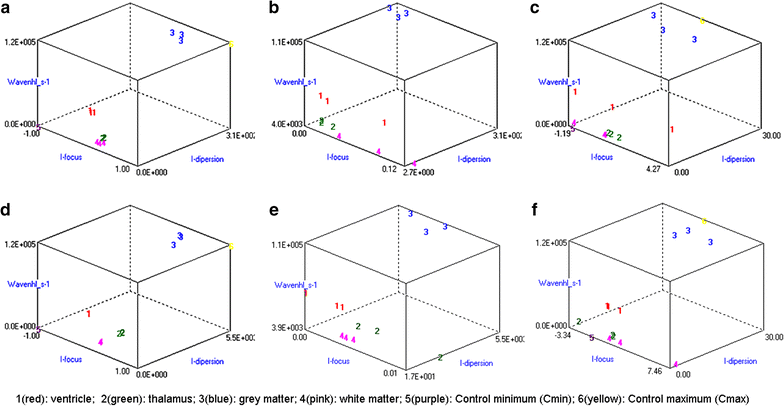
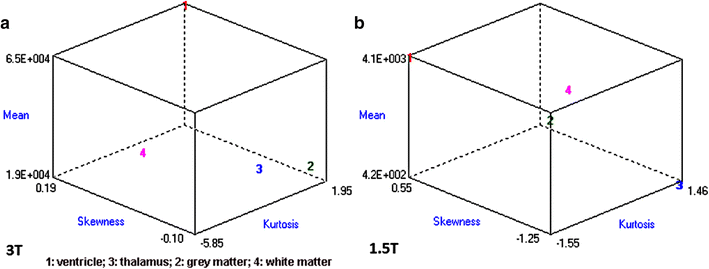
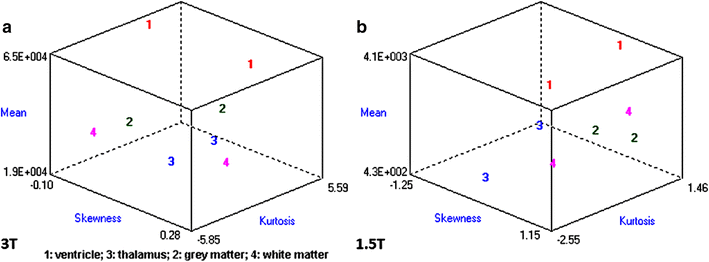
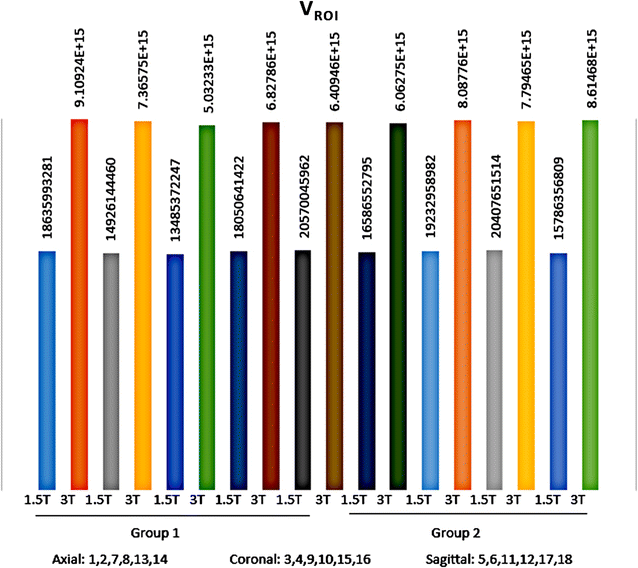
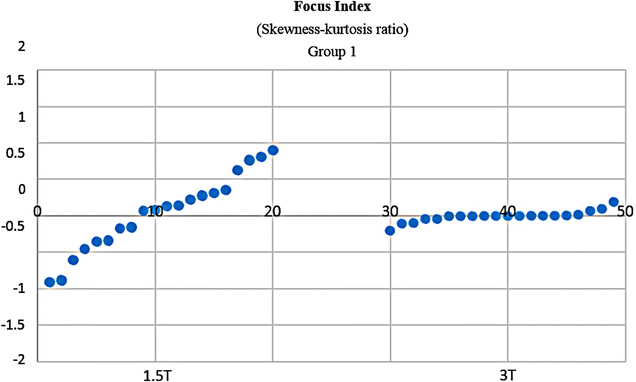
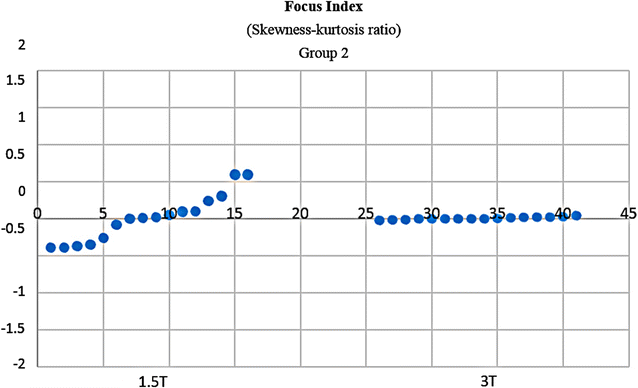
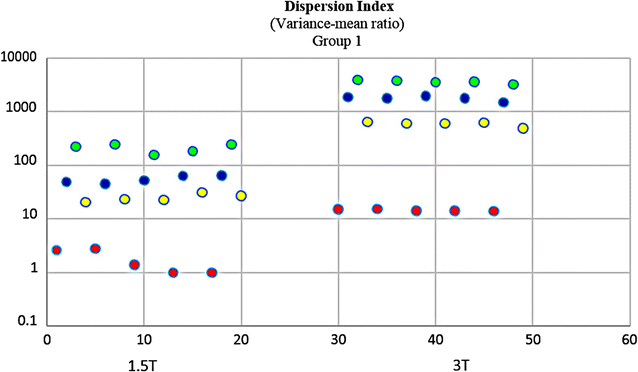
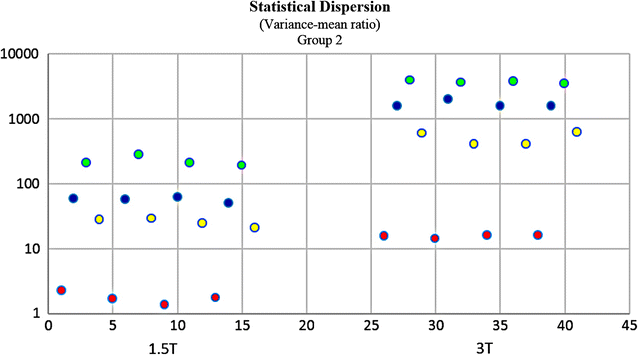
Results: Wavenhl, focus index, and dispersion index revealed better quality for 3 T. Though both 1.5 and 3 T images were 16-bit DICOM encoded, nearly 16 and 12 usable bits were measured in 3 and 1.5 T images, respectively. The four-bit padding observed in 1.5 T K-space encoding mimics noise by adding illusionistic details, which are not really part of the image. In contrast, zero-bit padding in 3 T provides space for storing more details and increases the likelihood of noise but as well as edges, which in turn are very crucial for differentiation of closely related anatomical structures.
Conclusions: Both encoding modes are possible with both units, but higher 3 T resolution is the main difference. It contributes to higher perceived and available dynamic range. Apart from surprisingly larger Fisher coefficient, no significant difference was observed when testing was conducted with down-converted 8-bit BMP images.
Keywords: Artificial intelligence; Computational visual cognition; Computer-assisted radiology; Fetal brain; Histogram; Hugues Gentillon; Mazda; Medical cybernetics; Prenatal development; Teleradiology; Wavelets; b11.
Figures
Similar articles
-
Improved medical image fusion based on cascaded PCA and shift invariant wavelet transforms.Int J Comput Assist Radiol Surg. 2018 Feb;13(2):229-240. doi: 10.1007/s11548-017-1692-4. Epub 2017 Dec 18. Int J Comput Assist Radiol Surg. 2018. PMID: 29250750
-
On the influence of zero-padding on the nonlinear operations in Quantitative Susceptibility Mapping.Magn Reson Imaging. 2017 Jan;35:154-159. doi: 10.1016/j.mri.2016.08.020. Epub 2016 Aug 29. Magn Reson Imaging. 2017. PMID: 27587225 Free PMC article.
-
CUDA-based acceleration and BPN-assisted automation of bilateral filtering for brain MR image restoration.Med Phys. 2017 Apr;44(4):1420-1436. doi: 10.1002/mp.12157. Med Phys. 2017. PMID: 28196280
-
Texture analysis of medical images.Clin Radiol. 2004 Dec;59(12):1061-9. doi: 10.1016/j.crad.2004.07.008. Clin Radiol. 2004. PMID: 15556588 Review.
-
Wavelets in temporal and spatial processing of biomedical images.Annu Rev Biomed Eng. 2000;2:511-50. doi: 10.1146/annurev.bioeng.2.1.511. Annu Rev Biomed Eng. 2000. PMID: 11701522 Review.
Cited by
-
Towards deep phenotyping pregnancy: a systematic review on artificial intelligence and machine learning methods to improve pregnancy outcomes.Brief Bioinform. 2021 Sep 2;22(5):bbaa369. doi: 10.1093/bib/bbaa369. Brief Bioinform. 2021. PMID: 33406530 Free PMC article.
-
Artificial intelligence applied to fetal MRI: A scoping review of current research.Br J Radiol. 2023 Jul;96(1147):20211205. doi: 10.1259/bjr.20211205. Epub 2022 Mar 18. Br J Radiol. 2023. PMID: 35286139 Free PMC article.
-
Texture analysis using short-tau inversion recovery magnetic resonance images to differentiate squamous cell carcinoma of the gingiva from medication-related osteonecrosis of the jaw.Oral Radiol. 2024 Apr;40(2):219-225. doi: 10.1007/s11282-023-00725-3. Epub 2023 Dec 7. Oral Radiol. 2024. PMID: 38060046
References
-
- Richards DS. Prenatal ultrasound to detect fetal anomalies. NeoReviews. 2012;13(1):e9–e19. doi: 10.1542/neo.13-1-e9. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

