Push-Pull and Feedback Mechanisms Can Align Signaling System Outputs with Inputs
- PMID: 27894998
- PMCID: PMC5134923
- DOI: 10.1016/j.cels.2016.10.002
Push-Pull and Feedback Mechanisms Can Align Signaling System Outputs with Inputs
Abstract
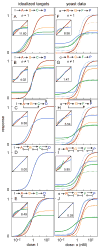
Many cell signaling systems, including the yeast pheromone response system, exhibit "dose-response alignment" (DoRA), in which output of one or more downstream steps closely matches the fraction of occupied receptors. DoRA can improve the fidelity of transmitted dose information. Here, we searched systematically for biochemical network topologies that produced DoRA. Most networks, including many containing feedback and feedforward loops, could not produce DoRA. However, networks including "push-pull" mechanisms, in which the active form of a signaling species stimulates downstream activity and the nominally inactive form reduces downstream activity, enabled perfect DoRA. Networks containing feedbacks enabled DoRA, but only if they also compared feedback to input and adjusted output to match. Our results establish push-pull as a non-feedback mechanism to align output with variable input and maximize information transfer in signaling systems. They also suggest genetic approaches to determine whether particular signaling systems use feedback or push-pull control.
Keywords: Saccharomyces cervisiae; cell signaling; dose response alignment; paradoxical signaling; pheromone response system; push-pull; yeast.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures
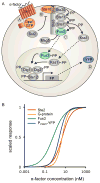
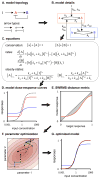
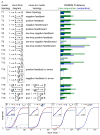
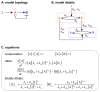


Comment in
-
Signaling Tug-of-War Delivers the Whole Message.Cell Syst. 2016 Nov 23;3(5):414-416. doi: 10.1016/j.cels.2016.11.003. Cell Syst. 2016. PMID: 27883887
References
-
- Alon U. Network motifs: theory and experimental approaches. Nature. 2007;8:450–461. - PubMed
-
- Ang J, Bagh S, Ingalls BP, McMillen DR. Considerations for using integral feedback control to construct a perfectly adapting synthetic gene network. J Theor Biol. 2010;266:723–738. - PubMed
-
- Astrom KJ, Murray RM. Feedback systems: an introduction for scientists and engineers. Princeton: Princeton University Press; 2008.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

