Functional exploration of colorectal cancer genomes using Drosophila
- PMID: 27897178
- PMCID: PMC5141297
- DOI: 10.1038/ncomms13615
Functional exploration of colorectal cancer genomes using Drosophila
Abstract
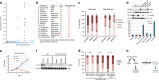
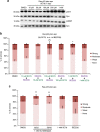
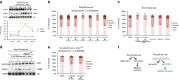
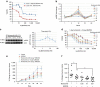
The multigenic nature of human tumours presents a fundamental challenge for cancer drug discovery. Here we use Drosophila to generate 32 multigenic models of colon cancer using patient data from The Cancer Genome Atlas. These models recapitulate key features of human cancer, often as emergent properties of multigenic combinations. Multigenic models such as ras p53 pten apc exhibit emergent resistance to a panel of cancer-relevant drugs. Exploring one drug in detail, we identify a mechanism of resistance for the PI3K pathway inhibitor BEZ235. We use this data to identify a combinatorial therapy that circumvents this resistance through a two-step process of emergent pathway dependence and sensitivity we term 'induced dependence'. This approach is effective in cultured human tumour cells, xenografts and mouse models of colorectal cancer. These data demonstrate how multigenic animal models that reference cancer genomes can provide an effective approach for developing novel targeted therapies.
Figures

Similar articles
-
Antitumor activity of pimasertib, a selective MEK 1/2 inhibitor, in combination with PI3K/mTOR inhibitors or with multi-targeted kinase inhibitors in pimasertib-resistant human lung and colorectal cancer cells.Int J Cancer. 2013 Nov;133(9):2089-101. doi: 10.1002/ijc.28236. Epub 2013 May 29. Int J Cancer. 2013. PMID: 23629727
-
Upregulation of microRNA-135b and microRNA-182 promotes chemoresistance of colorectal cancer by targeting ST6GALNAC2 via PI3K/AKT pathway.Mol Carcinog. 2017 Dec;56(12):2669-2680. doi: 10.1002/mc.22710. Epub 2017 Aug 21. Mol Carcinog. 2017. PMID: 28767179
-
Tankyrase Inhibition Blocks Wnt/β-Catenin Pathway and Reverts Resistance to PI3K and AKT Inhibitors in the Treatment of Colorectal Cancer.Clin Cancer Res. 2016 Feb 1;22(3):644-56. doi: 10.1158/1078-0432.CCR-14-3081. Epub 2015 Jul 29. Clin Cancer Res. 2016. PMID: 26224873
-
RES-529: a PI3K/AKT/mTOR pathway inhibitor that dissociates the mTORC1 and mTORC2 complexes.Anticancer Drugs. 2016 Jul;27(6):475-87. doi: 10.1097/CAD.0000000000000354. Anticancer Drugs. 2016. PMID: 26918392 Free PMC article. Review.
-
Targeting the translational apparatus to improve leukemia therapy: roles of the PI3K/PTEN/Akt/mTOR pathway.Leukemia. 2011 Jul;25(7):1064-79. doi: 10.1038/leu.2011.46. Epub 2011 Mar 25. Leukemia. 2011. PMID: 21436840 Review.
Cited by
-
Rational Design and Identification of Harmine-Inspired, N-Heterocyclic DYRK1A Inhibitors Employing a Functional Genomic In Vivo Drosophila Model System.ChemMedChem. 2022 Feb 16;17(4):e202100512. doi: 10.1002/cmdc.202100512. Epub 2022 Jan 27. ChemMedChem. 2022. PMID: 34994084 Free PMC article.
-
Drug screening in Drosophila; why, when, and when not?Wiley Interdiscip Rev Dev Biol. 2019 Nov;8(6):e346. doi: 10.1002/wdev.346. Epub 2019 May 5. Wiley Interdiscip Rev Dev Biol. 2019. PMID: 31056843 Free PMC article. Review.
-
Amalgam regulates the receptor tyrosine kinase pathway through Sprouty in glial cell development in the Drosophila larval brain.J Cell Sci. 2020 Oct 1;133(19):jcs250837. doi: 10.1242/jcs.250837. J Cell Sci. 2020. PMID: 32878945 Free PMC article.
-
Rounding up the Usual Suspects: Assessing Yorkie, AP-1, and Stat Coactivation in Tumorigenesis.Int J Mol Sci. 2020 Jun 27;21(13):4580. doi: 10.3390/ijms21134580. Int J Mol Sci. 2020. PMID: 32605129 Free PMC article.
-
A polycistronic transgene design for combinatorial genetic perturbations from a single transcript in Drosophila.PLoS Genet. 2023 Jun 2;19(6):e1010792. doi: 10.1371/journal.pgen.1010792. eCollection 2023 Jun. PLoS Genet. 2023. PMID: 37267433 Free PMC article.
References
-
- Stratton M. Exploring the genomes of cancer cells: progress and promise. Science 331, 1553–1558 (2011). - PubMed
-
- Biankin A. V., Piantadosi S. & Hollingsworth S. J. Patient-centric trials for therapeutic development in precision oncology. Nature 526, 361–370 (2015). - PubMed
-
- Mendelsohn J. Personalizing oncology: perspectives and prospects. J. Clin. Oncol. 31, 1904–1911 (2013). - PubMed
-
- Caponigro G. & Sellers W. R. Advances in the preclinical testing of cancer therapeutic hypotheses. Nat. Rev. Drug Discov. 10, 179–187 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

