Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases
- PMID: 27898009
- PMCID: PMC5187769
- DOI: 10.3390/ijms17121969
Effects of Glycosylation on the Enzymatic Activity and Mechanisms of Proteases
Abstract
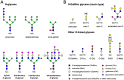
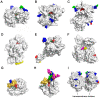
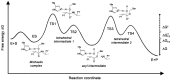
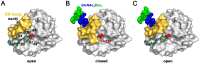
Posttranslational modifications are an important feature of most proteases in higher organisms, such as the conversion of inactive zymogens into active proteases. To date, little information is available on the role of glycosylation and functional implications for secreted proteases. Besides a stabilizing effect and protection against proteolysis, several proteases show a significant influence of glycosylation on the catalytic activity. Glycans can alter the substrate recognition, the specificity and binding affinity, as well as the turnover rates. However, there is currently no known general pattern, since glycosylation can have both stimulating and inhibiting effects on activity. Thus, a comparative analysis of individual cases with sufficient enzyme kinetic and structural data is a first approach to describe mechanistic principles that govern the effects of glycosylation on the function of proteases. The understanding of glycan functions becomes highly significant in proteomic and glycomic studies, which demonstrated that cancer-associated proteases, such as kallikrein-related peptidase 3, exhibit strongly altered glycosylation patterns in pathological cases. Such findings can contribute to a variety of future biomedical applications.
Keywords: Michaelis constant; N-glycosylation; O-glycosylation; core glycan; enzyme kinetics; flexible loops; secreted protease; sequon; substrate recognition; turnover number.
Conflict of interest statement
The author declares no conflict of interest. The founding sponsors had no role in the design of the study; in the analyses and interpretation of the literature; in the writing of the manuscript, and in the decision to publish the results.
Figures


References
-
- Varki A., Cummings R.D., Esko J.D., Freeze H.H., Stanley P., Bertozzi C.R., Hart G.W., Etzler M.E. In: Essentials of Glycobiology. 2nd ed. Varki A., Cummings R.D., Esko J.D., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2009. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

