Replication fork instability and the consequences of fork collisions from rereplication
- PMID: 27898391
- PMCID: PMC5110991
- DOI: 10.1101/gad.288142.116
Replication fork instability and the consequences of fork collisions from rereplication
Abstract
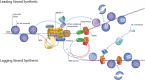
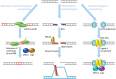
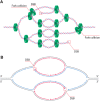
Replication forks encounter obstacles that must be repaired or bypassed to complete chromosome duplication before cell division. Proteomic analysis of replication forks suggests that the checkpoint and repair machinery travels with unperturbed forks, implying that they are poised to respond to stalling and collapse. However, impaired fork progression still generates aberrations, including repeat copy number instability and chromosome rearrangements. Deregulated origin firing also causes fork instability if a newer fork collides with an older one, generating double-strand breaks (DSBs) and partially rereplicated DNA. Current evidence suggests that multiple mechanisms are used to repair rereplication damage, yet these can have deleterious consequences for genome integrity.
Keywords: break-induced replication; double-strand break repair; gene amplification; homologous recombination; nonhomologous end-joining.
© 2016 Alexander and Orr-Weaver; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. 2000. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 60: 5934–5936. - PubMed
-
- Alabert C, Groth A. 2012. Chromatin replication and epigenome maintenance. Nat Rev Mol Cell Biol 13: 153–167. - PubMed
-
- Alabert C, Bukowski-Wills JC, Lee SB, Kustatscher G, Nakamura K, de Lima Alves F, Menard P, Mejlvang J, Rappsilber J, Groth A. 2014. Nascent chromatin capture proteomics determines chromatin dynamics during DNA replication and identifies unknown fork components. Nat Cell Biol 16: 281–293. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
