Comparative sequence analysis of Cyclospora cayetanensis apicoplast genomes originating from diverse geographical regions
- PMID: 27899155
- PMCID: PMC5129617
- DOI: 10.1186/s13071-016-1896-4
Comparative sequence analysis of Cyclospora cayetanensis apicoplast genomes originating from diverse geographical regions
Abstract
Background: Cyclospora cayetanensis is an emerging coccidian parasite that causes endemic and epidemic diarrheal disease called cyclosporiasis, and this infection is associated with consumption of contaminated produce or water in developed and developing regions. Food-borne outbreaks of cyclosporiasis have occurred almost every year in the USA since the 1990s. Investigations of these outbreaks are currently hampered due to lack of molecular epidemiological tools for trace back analysis. The apicoplast of C. cayetanensis, a relict non-photosynthetic plastid with an independent genome, provides an attractive target to discover sequence polymorphisms useful as genetic markers for detection and trace back analysis of the parasite. Distinct differences in the apicoplast genomes of C. cayetanensis could be useful in designing advanced molecular methods for rapid detection and, subtyping and geographical source attribution, which would aid outbreak investigations and surveillance studies.
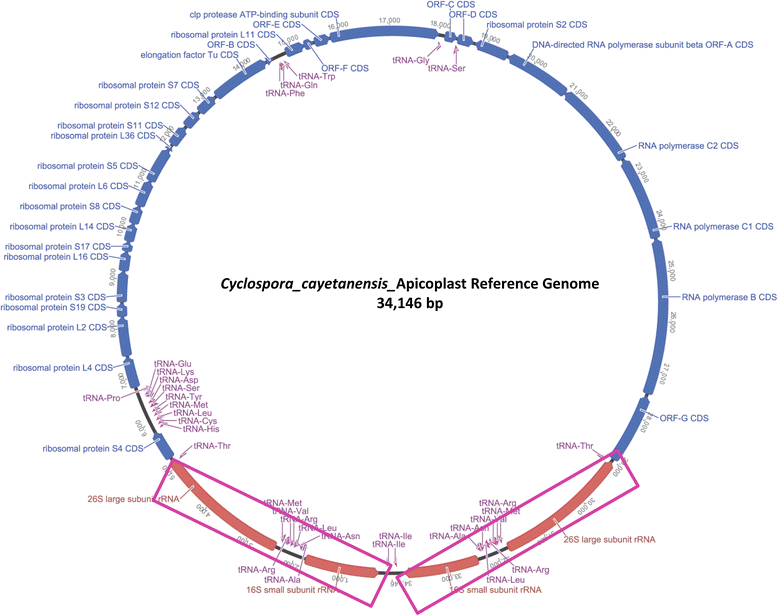
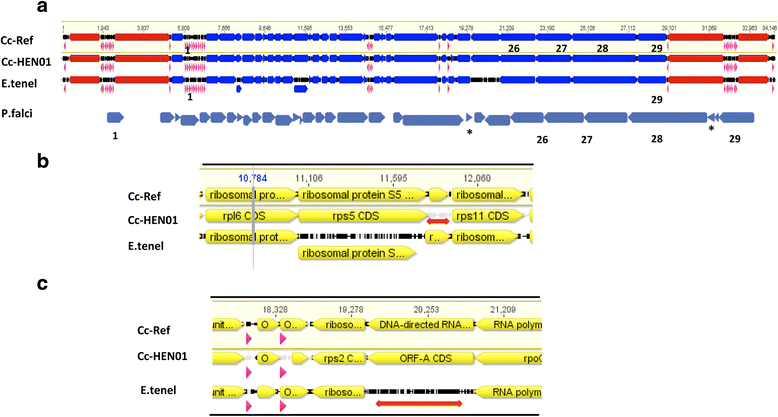
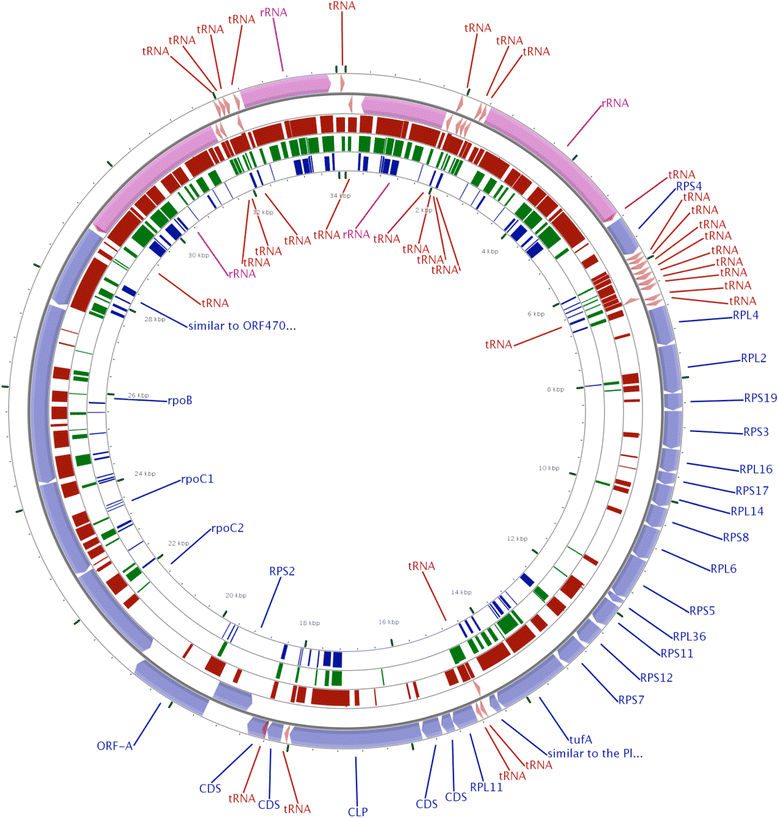
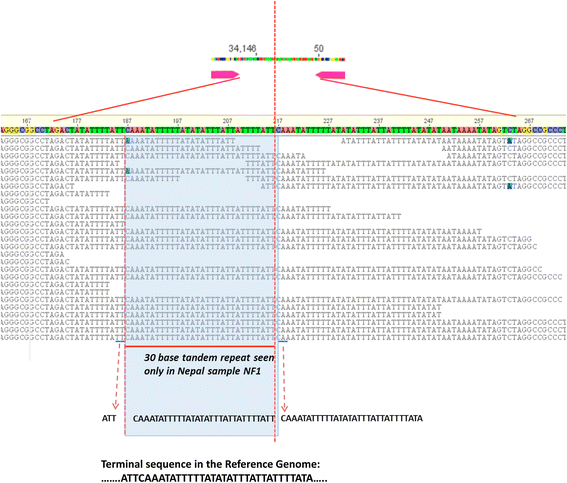
Methods: To obtain the genome sequence of the C. cayetanensis apicoplast, we sequenced the C. cayetanensis genomic DNA extracted from clinical stool samples, assembled and annotated a 34,146 bp-long circular sequence, and used this sequence as a reference genome in this study. We compared the genome and the predicted proteome to the data available from other apicomplexan parasites. To initialize the search for genetic markers, we mapped the raw sequence reads from an additional 11 distinct clinical stool samples originating from Nepal, New York, Texas, and Indonesia to the apicoplast reference genome.
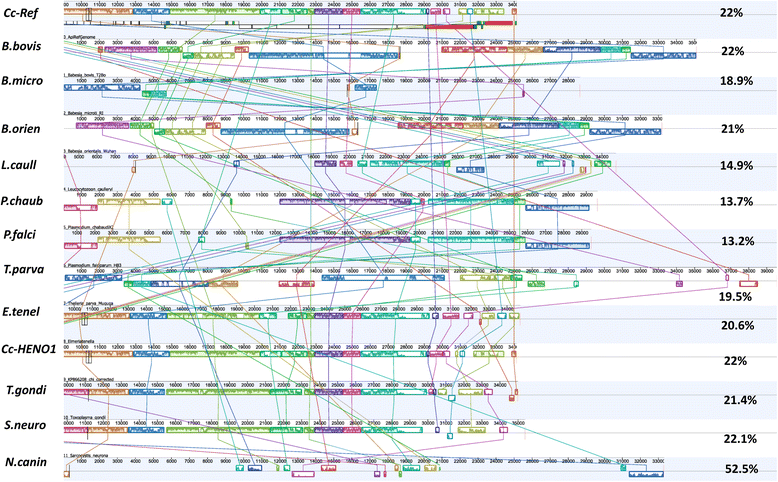
Results: We identified several high quality single nucleotide polymorphisms (SNPs) and small insertion/deletions spanning the apicoplast genome supported by extensive sequencing reads data, and a 30 bp sequence repeat at the terminal spacer region in a Nepalese sample. The predicted proteome consists of 29 core apicomplexan peptides found in most of the apicomplexans. Cluster analysis of these C. cayetanensis apicoplast genomes revealed a familiar pattern of tight grouping with Eimeria and Toxoplasma, separated from distant species such as Plasmodium and Babesia.
Conclusions: SNPs and sequence repeats identified in this study may be useful as genetic markers for identification and differentiation of C. cayetanensis isolates found and could facilitate outbreak investigations.
Keywords: Apicoplast genome; Cyclospora cayetanensis; Genomics; Next generation sequencing.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

