Bacillus subtilis Early Colonization of Arabidopsis thaliana Roots Involves Multiple Chemotaxis Receptors
- PMID: 27899502
- PMCID: PMC5137498
- DOI: 10.1128/mBio.01664-16
Bacillus subtilis Early Colonization of Arabidopsis thaliana Roots Involves Multiple Chemotaxis Receptors
Abstract
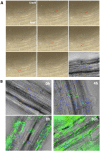
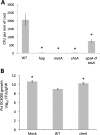
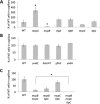
Colonization of plant roots by Bacillus subtilis is mutually beneficial to plants and bacteria. Plants can secrete up to 30% of their fixed carbon via root exudates, thereby feeding the bacteria, and in return the associated B. subtilis bacteria provide the plant with many growth-promoting traits. Formation of a biofilm on the root by matrix-producing B. subtilis is a well-established requirement for long-term colonization. However, we observed that cells start forming a biofilm only several hours after motile cells first settle on the plant. We also found that intact chemotaxis machinery is required for early root colonization by B. subtilis and for plant protection. Arabidopsis thaliana root exudates attract B. subtilis in vitro, an activity mediated by the two characterized chemoreceptors, McpB and McpC, as well as by the orphan receptor TlpC. Nonetheless, bacteria lacking these chemoreceptors are still able to colonize the root, suggesting that other chemoreceptors might also play a role in this process. These observations suggest that A. thaliana actively recruits B. subtilis through root-secreted molecules, and our results stress the important roles of B. subtilis chemoreceptors for efficient colonization of plants in natural environments. These results demonstrate a remarkable strategy adapted by beneficial rhizobacteria to utilize carbon-rich root exudates, which may facilitate rhizobacterial colonization and a mutualistic association with the host.
Importance: Bacillus subtilis is a plant growth-promoting rhizobacterium that establishes robust interactions with roots. Many studies have now demonstrated that biofilm formation is required for long-term colonization. However, we observed that motile B. subtilis mediates the first contact with the roots. These cells differentiate into biofilm-producing cells only several hours after the bacteria first contact the root. Our study reveals that intact chemotaxis machinery is required for the bacteria to reach the root. Many, if not all, of the B. subtilis 10 chemoreceptors are involved in the interaction with the plant. These observations stress the importance of root-bacterium interactions in the B. subtilis lifestyle.
Copyright © 2016 Allard-Massicotte et al.
Figures


References
-
- Gaworzewska ET, Carlile MJ. 1982. Positive chemotaxis of Rhizobium leguminosarum and other bacteria towards root exudates from legumes and other plants. Microbiology 128:1179–1188. doi:10.1099/00221287-128-6-1179. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

