The Zebrafish Model Organism Database: new support for human disease models, mutation details, gene expression phenotypes and searching
- PMID: 27899582
- PMCID: PMC5210580
- DOI: 10.1093/nar/gkw1116
The Zebrafish Model Organism Database: new support for human disease models, mutation details, gene expression phenotypes and searching
Abstract
The Zebrafish Model Organism Database (ZFIN; http://zfin.org) is the central resource for zebrafish (Danio rerio) genetic, genomic, phenotypic and developmental data. ZFIN curators provide expert manual curation and integration of comprehensive data involving zebrafish genes, mutants, transgenic constructs and lines, phenotypes, genotypes, gene expressions, morpholinos, TALENs, CRISPRs, antibodies, anatomical structures, models of human disease and publications. We integrate curated, directly submitted, and collaboratively generated data, making these available to zebrafish research community. Among the vertebrate model organisms, zebrafish are superbly suited for rapid generation of sequence-targeted mutant lines, characterization of phenotypes including gene expression patterns, and generation of human disease models. The recent rapid adoption of zebrafish as human disease models is making management of these data particularly important to both the research and clinical communities. Here, we describe recent enhancements to ZFIN including use of the zebrafish experimental conditions ontology, 'Fish' records in the ZFIN database, support for gene expression phenotypes, models of human disease, mutation details at the DNA, RNA and protein levels, and updates to the ZFIN single box search.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




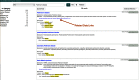
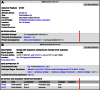
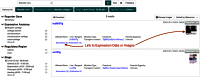
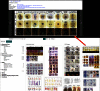
References
-
- Brush Matthew H, Mungall Chris J, Washington Nicole, Haendel M.A. ICBO 2013 - Proceedings of the 4th International Conference on Biomedical Ontology. Montreal: CEUR-WS; 2013. What's in a genotype?: an ontological characterization for integration of genetic variation data; pp. 105–108.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

