InterPro in 2017-beyond protein family and domain annotations
- PMID: 27899635
- PMCID: PMC5210578
- DOI: 10.1093/nar/gkw1107
InterPro in 2017-beyond protein family and domain annotations
Abstract
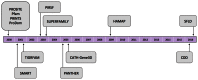
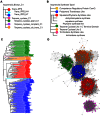
InterPro (http://www.ebi.ac.uk/interpro/) is a freely available database used to classify protein sequences into families and to predict the presence of important domains and sites. InterProScan is the underlying software that allows both protein and nucleic acid sequences to be searched against InterPro's predictive models, which are provided by its member databases. Here, we report recent developments with InterPro and its associated software, including the addition of two new databases (SFLD and CDD), and the functionality to include residue-level annotation and prediction of intrinsic disorder. These developments enrich the annotations provided by InterPro, increase the overall number of residues annotated and allow more specific functional inferences.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

