Open Targets: a platform for therapeutic target identification and validation
- PMID: 27899665
- PMCID: PMC5210543
- DOI: 10.1093/nar/gkw1055
Open Targets: a platform for therapeutic target identification and validation
Abstract
We have designed and developed a data integration and visualization platform that provides evidence about the association of known and potential drug targets with diseases. The platform is designed to support identification and prioritization of biological targets for follow-up. Each drug target is linked to a disease using integrated genome-wide data from a broad range of data sources. The platform provides either a target-centric workflow to identify diseases that may be associated with a specific target, or a disease-centric workflow to identify targets that may be associated with a specific disease. Users can easily transition between these target- and disease-centric workflows. The Open Targets Validation Platform is accessible at https://www.targetvalidation.org.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

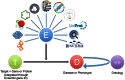
References
-
- Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., Pangalos M.N. Lessons learned from the fate of AstraZeneca's drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 2014;13:419–431. - PubMed
-
- Arrowsmith J., Miller P. Trial watch: phase II and phase III attrition rates 2011-2012. Nat. Rev. Drug Discov. 2013;12:569. - PubMed
-
- Plenge R.M. Disciplined approach to drug discovery and early development. Sci. Transl. Med. 2016;8:349ps315. - PubMed

