The mouse Gene Expression Database (GXD): 2017 update
- PMID: 27899677
- PMCID: PMC5210556
- DOI: 10.1093/nar/gkw1073
The mouse Gene Expression Database (GXD): 2017 update
Abstract
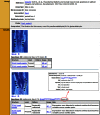
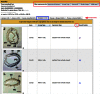
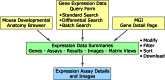
The Gene Expression Database (GXD; www.informatics.jax.org/expression.shtml) is an extensive and well-curated community resource of mouse developmental expression information. Through curation of the scientific literature and by collaborations with large-scale expression projects, GXD collects and integrates data from RNA in situ hybridization, immunohistochemistry, RT-PCR, northern blot and western blot experiments. Expression data from both wild-type and mutant mice are included. The expression data are combined with genetic and phenotypic data in Mouse Genome Informatics (MGI) and made readily accessible to many types of database searches. At present, GXD includes over 1.5 million expression results and more than 300 000 images, all annotated with detailed and standardized metadata. Since our last report in 2014, we have added a large amount of data, we have enhanced data and database infrastructure, and we have implemented many new search and display features. Interface enhancements include: a new Mouse Developmental Anatomy Browser; interactive tissue-by-developmental stage and tissue-by-gene matrix views; capabilities to filter and sort expression data summaries; a batch search utility; gene-based expression overviews; and links to expression data from other species.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Ringwald M., Baldock R., Bard J., Kaufman M., Eppig J.T., Richardson J.E., Nadeau J.H., Davidson D. A database for mouse development. Science. 1994;265:2033–2034. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

