Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells
- PMID: 27901044
- PMCID: PMC5141517
- DOI: 10.1038/ncomms13683
Fatty acid metabolic reprogramming via mTOR-mediated inductions of PPARγ directs early activation of T cells
Abstract
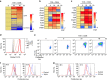
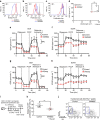
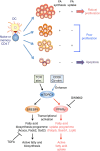
To fulfil the bioenergetic requirements for increased cell size and clonal expansion, activated T cells reprogramme their metabolic signatures from energetically quiescent to activated. However, the molecular mechanisms and essential components controlling metabolic reprogramming in T cells are not well understood. Here, we show that the mTORC1-PPARγ pathway is crucial for the fatty acid uptake programme in activated CD4+ T cells. This pathway is required for full activation and rapid proliferation of naive and memory CD4+ T cells. PPARγ directly binds and induces genes associated with fatty acid uptake in CD4+ T cells in both mice and humans. The PPARγ-dependent fatty acid uptake programme is critical for metabolic reprogramming. Thus, we provide important mechanistic insights into the metabolic reprogramming mechanisms that govern the expression of key enzymes, fatty acid metabolism and the acquisition of an activated phenotype during CD4+ T cell activation.
Figures





References
-
- Nakayama T. & Yamashita M. The TCR-mediated signaling pathways that control the direction of helper T cell differentiation. Semin. Immunol. 22, 303–309 (2010). - PubMed
-
- Colombetti S., Basso V., Mueller D. L. & Mondino A. Prolonged TCR/CD28 engagement drives IL-2-independent T cell clonal expansion through signaling mediated by the mammalian target of rapamycin. J. Immunol. 176, 2730–2738 (2006). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

