Structural basis for the disaggregase activity and regulation of Hsp104
- PMID: 27901467
- PMCID: PMC5130295
- DOI: 10.7554/eLife.21516
Structural basis for the disaggregase activity and regulation of Hsp104
Abstract
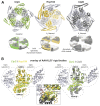
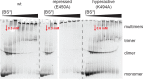
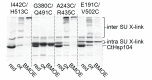
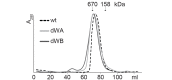
The Hsp104 disaggregase is a two-ring ATPase machine that rescues various forms of non-native proteins including the highly resistant amyloid fibers. The structural-mechanistic underpinnings of how the recovery of toxic protein aggregates is promoted and how this potent unfolding activity is prevented from doing collateral damage to cellular proteins are not well understood. Here, we present structural and biochemical data revealing the organization of Hsp104 from Chaetomium thermophilum at 3.7 Å resolution. We show that the coiled-coil domains encircling the disaggregase constitute a 'restraint mask' that sterically controls the mobility and thus the unfolding activity of the ATPase modules. In addition, we identify a mechanical linkage that coordinates the activity of the two ATPase rings and accounts for the high unfolding potential of Hsp104. Based on these findings, we propose a general model for how Hsp104 and related chaperones operate and are kept under control until recruited to appropriate substrates.
Keywords: C. thermophilum,; Protein disaggregase; Structual biology; biochemistry; biophysics; none; structural biology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures













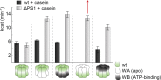


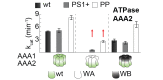

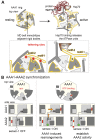


Similar articles
-
Split conformation of Chaetomium thermophilum Hsp104 disaggregase.Structure. 2021 Jul 1;29(7):721-730.e6. doi: 10.1016/j.str.2021.02.002. Epub 2021 Mar 1. Structure. 2021. PMID: 33651974
-
Structural determinants for protein unfolding and translocation by the Hsp104 protein disaggregase.Biosci Rep. 2017 Dec 22;37(6):BSR20171399. doi: 10.1042/BSR20171399. Print 2017 Dec 22. Biosci Rep. 2017. PMID: 29175998 Free PMC article.
-
Selective Hsp70-Dependent Docking of Hsp104 to Protein Aggregates Protects the Cell from the Toxicity of the Disaggregase.J Mol Biol. 2019 May 17;431(11):2180-2196. doi: 10.1016/j.jmb.2019.04.014. Epub 2019 Apr 24. J Mol Biol. 2019. PMID: 31026451
-
Mechanistic and Structural Insights into the Prion-Disaggregase Activity of Hsp104.J Mol Biol. 2016 May 8;428(9 Pt B):1870-85. doi: 10.1016/j.jmb.2015.11.016. Epub 2015 Dec 1. J Mol Biol. 2016. PMID: 26608812 Free PMC article. Review.
-
Chaperone-assisted protein aggregate reactivation: Different solutions for the same problem.Arch Biochem Biophys. 2015 Aug 15;580:121-34. doi: 10.1016/j.abb.2015.07.006. Epub 2015 Jul 6. Arch Biochem Biophys. 2015. PMID: 26159839 Review.
Cited by
-
Dynamic structural states of ClpB involved in its disaggregation function.Nat Commun. 2018 Jun 1;9(1):2147. doi: 10.1038/s41467-018-04587-w. Nat Commun. 2018. PMID: 29858573 Free PMC article.
-
Substrate Discrimination by ClpB and Hsp104.Front Mol Biosci. 2017 May 29;4:36. doi: 10.3389/fmolb.2017.00036. eCollection 2017. Front Mol Biosci. 2017. PMID: 28611991 Free PMC article.
-
Factors underlying asymmetric pore dynamics of disaggregase and microtubule-severing AAA+ machines.Biophys J. 2021 Aug 17;120(16):3437-3454. doi: 10.1016/j.bpj.2021.05.027. Epub 2021 Jun 25. Biophys J. 2021. PMID: 34181904 Free PMC article.
-
Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.Sci Adv. 2020 May 20;6(21):eaba8404. doi: 10.1126/sciadv.aba8404. eCollection 2020 May. Sci Adv. 2020. PMID: 32490208 Free PMC article.
-
Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.Science. 2017 Jul 21;357(6348):273-279. doi: 10.1126/science.aan1052. Epub 2017 Jun 15. Science. 2017. PMID: 28619716 Free PMC article.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallographica. Section D, Biological Crystallography. 1998;54:905–921. doi: 10.1107/s0907444998003254. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

